Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
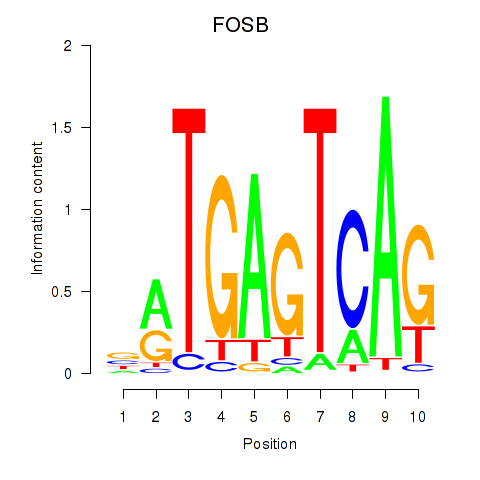
Results for FOSB
Z-value: 0.97
Transcription factors associated with FOSB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSB
|
ENSG00000125740.9 | FosB proto-oncogene, AP-1 transcription factor subunit |
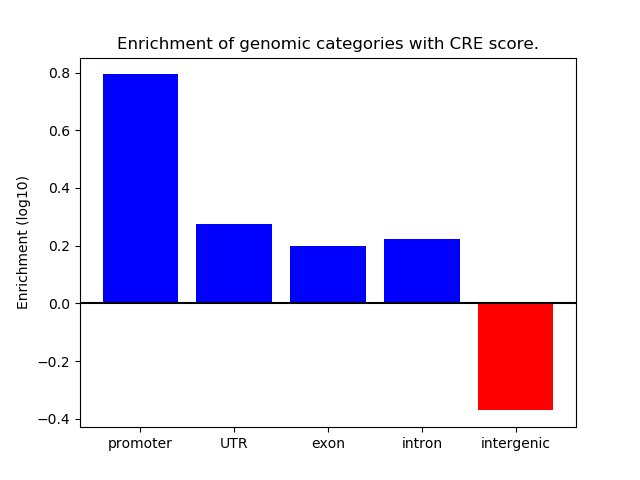
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_45970997_45971195 | FOSB | 157 | 0.919518 | -0.62 | 7.3e-02 | Click! |
| chr19_45973015_45973567 | FOSB | 120 | 0.935112 | -0.48 | 2.0e-01 | Click! |
| chr19_45975714_45975883 | FOSB | 2275 | 0.177319 | 0.43 | 2.5e-01 | Click! |
| chr19_45975959_45976300 | FOSB | 2606 | 0.160559 | 0.38 | 3.1e-01 | Click! |
| chr19_45973955_45974106 | FOSB | 507 | 0.666266 | -0.36 | 3.5e-01 | Click! |
Activity of the FOSB motif across conditions
Conditions sorted by the z-value of the FOSB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
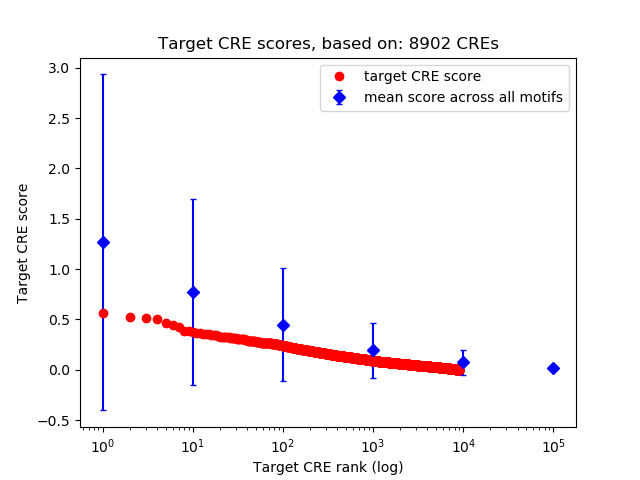
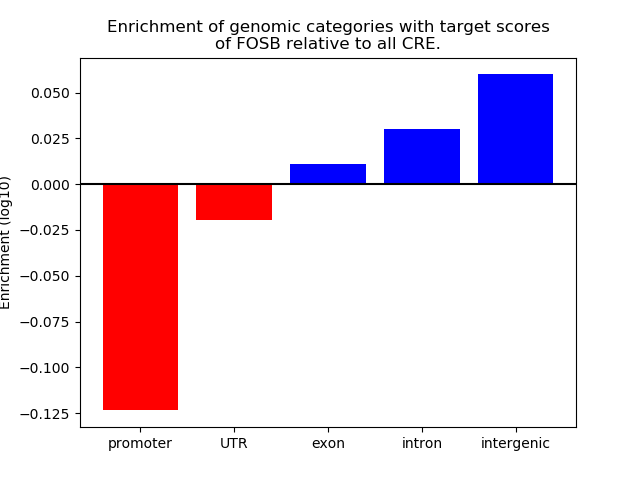
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_131593477_131594185 | 0.57 |
PDLIM4 |
PDZ and LIM domain 4 |
431 |
0.81 |
| chr1_44513542_44514157 | 0.53 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
16715 |
0.14 |
| chr12_72667295_72667865 | 0.51 |
TRHDE |
thyrotropin-releasing hormone degrading enzyme |
317 |
0.85 |
| chr20_30328267_30328442 | 0.51 |
TPX2 |
TPX2, microtubule-associated |
1280 |
0.36 |
| chr4_107956832_107957922 | 0.46 |
DKK2 |
dickkopf WNT signaling pathway inhibitor 2 |
77 |
0.99 |
| chr7_32109441_32109701 | 0.44 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
890 |
0.75 |
| chr7_77169254_77169538 | 0.43 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
2010 |
0.44 |
| chr4_141217669_141218059 | 0.39 |
ENSG00000252300 |
. |
21598 |
0.19 |
| chr5_158611818_158612659 | 0.39 |
RNF145 |
ring finger protein 145 |
22404 |
0.16 |
| chr13_40683698_40683888 | 0.38 |
ENSG00000207458 |
. |
117171 |
0.07 |
| chr13_103478539_103479046 | 0.37 |
ENSG00000222301 |
. |
6449 |
0.13 |
| chr5_15620667_15620944 | 0.37 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
4714 |
0.28 |
| chr17_9018882_9019405 | 0.36 |
NTN1 |
netrin 1 |
47109 |
0.15 |
| chr4_77506894_77507309 | 0.36 |
ENSG00000265314 |
. |
10397 |
0.17 |
| chr11_118478413_118479735 | 0.35 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
716 |
0.54 |
| chr2_202774749_202774932 | 0.35 |
ENSG00000212184 |
. |
17322 |
0.18 |
| chr4_71587677_71588145 | 0.34 |
RUFY3 |
RUN and FYVE domain containing 3 |
183 |
0.92 |
| chr17_65427494_65427948 | 0.34 |
ENSG00000201547 |
. |
22724 |
0.12 |
| chr3_99359127_99359366 | 0.33 |
COL8A1 |
collagen, type VIII, alpha 1 |
1792 |
0.46 |
| chr18_9743562_9744068 | 0.33 |
RAB31 |
RAB31, member RAS oncogene family |
35653 |
0.16 |
| chr3_159483400_159483608 | 0.33 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
612 |
0.7 |
| chr19_48928010_48928416 | 0.33 |
GRWD1 |
glutamate-rich WD repeat containing 1 |
20817 |
0.09 |
| chr6_169828167_169828442 | 0.33 |
THBS2 |
thrombospondin 2 |
174165 |
0.03 |
| chr11_10920623_10921193 | 0.32 |
ZBED5-AS1 |
ZBED5 antisense RNA 1 |
34100 |
0.15 |
| chr2_167230570_167230946 | 0.32 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
1739 |
0.43 |
| chr12_58129779_58130334 | 0.32 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
1563 |
0.16 |
| chr5_14268280_14268580 | 0.32 |
TRIO |
trio Rho guanine nucleotide exchange factor |
22656 |
0.28 |
| chr17_74442541_74443075 | 0.32 |
UBE2O |
ubiquitin-conjugating enzyme E2O |
6480 |
0.12 |
| chr8_23399124_23399403 | 0.32 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
12690 |
0.16 |
| chr3_53303838_53304218 | 0.31 |
ENSG00000238565 |
. |
13291 |
0.12 |
| chr4_23890131_23890306 | 0.31 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
1440 |
0.57 |
| chr19_47735595_47736220 | 0.31 |
BBC3 |
BCL2 binding component 3 |
116 |
0.95 |
| chr9_21677527_21678485 | 0.31 |
ENSG00000244230 |
. |
21307 |
0.21 |
| chr12_29310273_29310492 | 0.31 |
FAR2 |
fatty acyl CoA reductase 2 |
8249 |
0.26 |
| chr8_132828485_132828822 | 0.31 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
87682 |
0.1 |
| chr2_69059738_69059974 | 0.31 |
AC097495.2 |
|
680 |
0.71 |
| chr9_124044297_124044511 | 0.31 |
RP11-477J21.6 |
|
604 |
0.65 |
| chr12_112204695_112205567 | 0.30 |
ALDH2 |
aldehyde dehydrogenase 2 family (mitochondrial) |
405 |
0.84 |
| chr3_172280513_172281223 | 0.29 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
39571 |
0.16 |
| chr11_57241521_57241817 | 0.29 |
RP11-624G17.3 |
|
3338 |
0.12 |
| chr20_2367570_2367721 | 0.29 |
TGM6 |
transglutaminase 6 |
6091 |
0.22 |
| chr18_47250302_47250457 | 0.29 |
ACAA2 |
acetyl-CoA acyltransferase 2 |
87522 |
0.06 |
| chr3_105663549_105663788 | 0.29 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
75272 |
0.13 |
| chr1_221209086_221209419 | 0.29 |
HLX |
H2.0-like homeobox |
154668 |
0.04 |
| chr10_95361036_95361311 | 0.29 |
RBP4 |
retinol binding protein 4, plasma |
190 |
0.93 |
| chr2_220173306_220173592 | 0.29 |
PTPRN |
protein tyrosine phosphatase, receptor type, N |
257 |
0.82 |
| chr8_6404923_6405335 | 0.29 |
ANGPT2 |
angiopoietin 2 |
15436 |
0.25 |
| chr8_52782524_52782885 | 0.28 |
PCMTD1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
24399 |
0.15 |
| chr1_156863527_156863998 | 0.28 |
PEAR1 |
platelet endothelial aggregation receptor 1 |
239 |
0.89 |
| chr3_159745160_159745451 | 0.28 |
LINC01100 |
long intergenic non-protein coding RNA 1100 |
11494 |
0.19 |
| chr3_138581962_138582285 | 0.28 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
28343 |
0.2 |
| chr6_52217103_52217563 | 0.28 |
PAQR8 |
progestin and adipoQ receptor family member VIII |
8886 |
0.23 |
| chr16_80933820_80933971 | 0.28 |
CDYL2 |
chromodomain protein, Y-like 2 |
95669 |
0.06 |
| chr12_56075239_56075869 | 0.28 |
METTL7B |
methyltransferase like 7B |
224 |
0.86 |
| chr19_19574756_19575086 | 0.27 |
GATAD2A |
GATA zinc finger domain containing 2A |
570 |
0.67 |
| chr11_35641995_35642146 | 0.27 |
FJX1 |
four jointed box 1 (Drosophila) |
2335 |
0.36 |
| chr11_63273553_63274069 | 0.27 |
LGALS12 |
lectin, galactoside-binding, soluble, 12 |
24 |
0.97 |
| chr8_22343066_22343217 | 0.27 |
PPP3CC |
protein phosphatase 3, catalytic subunit, gamma isozyme |
10036 |
0.15 |
| chr1_233249017_233249707 | 0.27 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
46691 |
0.19 |
| chr11_12108503_12108884 | 0.27 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
6850 |
0.29 |
| chr13_107001311_107002028 | 0.27 |
EFNB2 |
ephrin-B2 |
185793 |
0.03 |
| chr4_71570546_71572213 | 0.27 |
RUFY3 |
RUN and FYVE domain containing 3 |
861 |
0.36 |
| chrX_134124848_134125476 | 0.27 |
SMIM10 |
small integral membrane protein 10 |
194 |
0.94 |
| chr7_47493101_47493387 | 0.27 |
TNS3 |
tensin 3 |
415 |
0.92 |
| chr20_20714689_20714840 | 0.27 |
ENSG00000264361 |
. |
4610 |
0.29 |
| chr4_159090784_159091060 | 0.27 |
FAM198B |
family with sequence similarity 198, member B |
608 |
0.57 |
| chr3_186470882_186471297 | 0.26 |
RP11-573D15.8 |
|
164 |
0.89 |
| chr5_33891538_33892159 | 0.26 |
ADAMTS12 |
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
198 |
0.94 |
| chr2_37116896_37117561 | 0.26 |
ENSG00000252756 |
. |
22590 |
0.18 |
| chr2_28565345_28565858 | 0.26 |
AC093690.1 |
|
32275 |
0.15 |
| chr18_47016915_47017206 | 0.26 |
ENSG00000206602 |
. |
657 |
0.29 |
| chr3_25614345_25614537 | 0.26 |
ENSG00000272166 |
. |
17344 |
0.22 |
| chr3_151046365_151046540 | 0.26 |
P2RY13 |
purinergic receptor P2Y, G-protein coupled, 13 |
884 |
0.57 |
| chr18_60051802_60052141 | 0.26 |
RP11-640A1.3 |
|
6608 |
0.23 |
| chr6_7730907_7731182 | 0.26 |
BMP6 |
bone morphogenetic protein 6 |
4014 |
0.34 |
| chr3_146221452_146221714 | 0.26 |
PLSCR2 |
phospholipid scramblase 2 |
7805 |
0.22 |
| chr4_138451566_138451717 | 0.26 |
PCDH18 |
protocadherin 18 |
1924 |
0.52 |
| chr20_44443275_44443426 | 0.26 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
1705 |
0.18 |
| chr12_77257769_77258091 | 0.26 |
RP11-461F16.3 |
|
14043 |
0.22 |
| chr11_125497290_125497441 | 0.26 |
CHEK1 |
checkpoint kinase 1 |
722 |
0.57 |
| chr12_8043530_8043933 | 0.26 |
SLC2A14 |
solute carrier family 2 (facilitated glucose transporter), member 14 |
13 |
0.96 |
| chr15_26332761_26333025 | 0.25 |
ENSG00000212604 |
. |
78672 |
0.11 |
| chr4_73433751_73434036 | 0.25 |
ADAMTS3 |
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
623 |
0.85 |
| chr1_180126237_180126440 | 0.25 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
2306 |
0.36 |
| chr1_231297648_231298418 | 0.25 |
TRIM67 |
tripartite motif containing 67 |
175 |
0.95 |
| chr2_18157972_18158481 | 0.25 |
ENSG00000212455 |
. |
63773 |
0.13 |
| chr8_71359441_71359951 | 0.25 |
ENSG00000253143 |
. |
33026 |
0.17 |
| chr17_55980324_55980725 | 0.25 |
CUEDC1 |
CUE domain containing 1 |
226 |
0.93 |
| chr11_94829276_94829597 | 0.25 |
ENDOD1 |
endonuclease domain containing 1 |
6462 |
0.24 |
| chr15_75943747_75944181 | 0.25 |
SNX33 |
sorting nexin 33 |
1867 |
0.18 |
| chr10_114745695_114746021 | 0.25 |
RP11-57H14.2 |
|
34224 |
0.18 |
| chr15_80452796_80453143 | 0.25 |
FAH |
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
4676 |
0.25 |
| chr12_113859774_113860410 | 0.25 |
SDSL |
serine dehydratase-like |
50 |
0.97 |
| chr4_138975164_138975441 | 0.24 |
ENSG00000250033 |
. |
3316 |
0.4 |
| chr20_50063597_50063893 | 0.24 |
ENSG00000266761 |
. |
5769 |
0.29 |
| chr1_17866743_17867038 | 0.24 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
560 |
0.82 |
| chr17_26220981_26221377 | 0.24 |
LYRM9 |
LYR motif containing 9 |
770 |
0.4 |
| chr7_40509791_40509942 | 0.24 |
AC004988.1 |
|
76661 |
0.12 |
| chr10_74572337_74572942 | 0.24 |
RP11-354E23.5 |
|
46321 |
0.13 |
| chr2_238768253_238768964 | 0.24 |
RAMP1 |
receptor (G protein-coupled) activity modifying protein 1 |
11 |
0.98 |
| chr7_21981069_21981345 | 0.24 |
CDCA7L |
cell division cycle associated 7-like |
3284 |
0.37 |
| chr2_200844549_200844884 | 0.24 |
C2orf47 |
chromosome 2 open reading frame 47 |
24172 |
0.18 |
| chr9_129884150_129884930 | 0.24 |
ANGPTL2 |
angiopoietin-like 2 |
373 |
0.9 |
| chr9_100639076_100639372 | 0.24 |
FOXE1 |
forkhead box E1 (thyroid transcription factor 2) |
23688 |
0.15 |
| chr18_416158_416713 | 0.24 |
RP11-720L2.2 |
|
7981 |
0.25 |
| chr2_114643988_114644188 | 0.23 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
3449 |
0.28 |
| chr1_211055833_211056141 | 0.23 |
KCNH1-IT1 |
KCNH1 intronic transcript 1 (non-protein coding) |
250729 |
0.02 |
| chr11_1770006_1770588 | 0.23 |
IFITM10 |
interferon induced transmembrane protein 10 |
1524 |
0.22 |
| chr3_12549443_12549677 | 0.23 |
ENSG00000200114 |
. |
3152 |
0.17 |
| chr9_3508149_3508375 | 0.23 |
RFX3 |
regulatory factor X, 3 (influences HLA class II expression) |
17721 |
0.25 |
| chr3_11571277_11571505 | 0.23 |
VGLL4 |
vestigial like 4 (Drosophila) |
39007 |
0.18 |
| chr19_38422631_38423016 | 0.23 |
SIPA1L3 |
signal-induced proliferation-associated 1 like 3 |
24955 |
0.15 |
| chr9_97663249_97663511 | 0.23 |
RP11-49O14.2 |
|
669 |
0.72 |
| chr3_128201686_128201886 | 0.23 |
GATA2 |
GATA binding protein 2 |
4973 |
0.17 |
| chr17_33378351_33378574 | 0.23 |
ENSG00000238858 |
. |
3051 |
0.16 |
| chr7_120723258_120723409 | 0.23 |
ENSG00000212628 |
. |
534 |
0.81 |
| chr5_67629922_67630135 | 0.23 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
41632 |
0.21 |
| chr1_31501484_31501723 | 0.23 |
PUM1 |
pumilio RNA-binding family member 1 |
33160 |
0.16 |
| chr19_6087775_6088269 | 0.23 |
CTC-232P5.3 |
|
20058 |
0.13 |
| chr5_141740462_141740706 | 0.23 |
AC005592.2 |
|
7476 |
0.26 |
| chr11_78885167_78885414 | 0.22 |
TENM4 |
teneurin transmembrane protein 4 |
41664 |
0.21 |
| chr1_32043259_32043540 | 0.22 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
1260 |
0.35 |
| chrX_114796055_114796359 | 0.22 |
PLS3 |
plastin 3 |
625 |
0.52 |
| chr9_29039310_29039461 | 0.22 |
ENSG00000215939 |
. |
150432 |
0.05 |
| chr17_66755661_66755820 | 0.22 |
ENSG00000263690 |
. |
6960 |
0.3 |
| chr6_76059036_76059425 | 0.22 |
RP11-415D17.3 |
|
7700 |
0.18 |
| chr18_3037544_3037723 | 0.22 |
ENSG00000252258 |
. |
12069 |
0.17 |
| chr17_1666268_1666531 | 0.22 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
276 |
0.85 |
| chr3_5181342_5181587 | 0.22 |
ARL8B |
ADP-ribosylation factor-like 8B |
17483 |
0.16 |
| chr6_162052900_162053180 | 0.22 |
ENSG00000221021 |
. |
16942 |
0.31 |
| chr8_126556861_126557105 | 0.22 |
ENSG00000266452 |
. |
100176 |
0.08 |
| chr17_13501346_13501683 | 0.22 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
3730 |
0.32 |
| chr10_6763782_6764322 | 0.22 |
PRKCQ |
protein kinase C, theta |
141789 |
0.05 |
| chr10_99474521_99475206 | 0.22 |
MARVELD1 |
MARVEL domain containing 1 |
1382 |
0.36 |
| chr1_192873657_192873883 | 0.21 |
ENSG00000223075 |
. |
28655 |
0.21 |
| chr9_89049595_89049782 | 0.21 |
ENSG00000222293 |
. |
11645 |
0.28 |
| chr5_98396547_98396924 | 0.21 |
ENSG00000200351 |
. |
124284 |
0.06 |
| chr3_188599058_188599349 | 0.21 |
TPRG1 |
tumor protein p63 regulated 1 |
65800 |
0.14 |
| chr9_12859535_12860149 | 0.21 |
ENSG00000222658 |
. |
25478 |
0.2 |
| chr1_68024994_68025388 | 0.21 |
ENSG00000207504 |
. |
18381 |
0.23 |
| chr6_113198454_113198633 | 0.21 |
ENSG00000201386 |
. |
94152 |
0.09 |
| chr4_119839531_119839737 | 0.21 |
SYNPO2 |
synaptopodin 2 |
29442 |
0.23 |
| chr4_99298536_99298978 | 0.21 |
RAP1GDS1 |
RAP1, GTP-GDP dissociation stimulator 1 |
1496 |
0.54 |
| chr8_16689434_16690090 | 0.21 |
ENSG00000264092 |
. |
31243 |
0.24 |
| chr8_104154291_104154696 | 0.21 |
C8orf56 |
chromosome 8 open reading frame 56 |
790 |
0.45 |
| chr12_64565605_64565889 | 0.21 |
RP11-196H14.3 |
|
24110 |
0.14 |
| chr16_55787112_55787263 | 0.21 |
CES1P1 |
carboxylesterase 1 pseudogene 1 |
7273 |
0.24 |
| chr2_197852750_197853145 | 0.21 |
ANKRD44 |
ankyrin repeat domain 44 |
12279 |
0.25 |
| chr6_167002477_167002628 | 0.21 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
38173 |
0.16 |
| chr14_106379215_106379366 | 0.21 |
ENSG00000233655 |
. |
194 |
0.61 |
| chr2_158117981_158118132 | 0.21 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
3946 |
0.28 |
| chr15_52860024_52861356 | 0.21 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
339 |
0.87 |
| chr9_111773050_111773591 | 0.21 |
CTNNAL1 |
catenin (cadherin-associated protein), alpha-like 1 |
2459 |
0.23 |
| chr9_130856416_130856740 | 0.21 |
RP11-379C10.4 |
|
158 |
0.89 |
| chr11_71811615_71811947 | 0.21 |
LAMTOR1 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
1477 |
0.2 |
| chr16_85548258_85548499 | 0.21 |
ENSG00000264203 |
. |
73280 |
0.1 |
| chr8_108509260_108510202 | 0.21 |
ANGPT1 |
angiopoietin 1 |
507 |
0.89 |
| chr3_185463378_185463630 | 0.21 |
ENSG00000265470 |
. |
22188 |
0.19 |
| chr2_232476974_232477319 | 0.20 |
C2orf57 |
chromosome 2 open reading frame 57 |
19571 |
0.16 |
| chr5_71643973_71644124 | 0.20 |
PTCD2 |
pentatricopeptide repeat domain 2 |
27823 |
0.16 |
| chr5_87441398_87441596 | 0.20 |
TMEM161B |
transmembrane protein 161B |
74951 |
0.11 |
| chr8_26123485_26124097 | 0.20 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
25216 |
0.23 |
| chr5_14036568_14037099 | 0.20 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
92181 |
0.09 |
| chr12_132221740_132221934 | 0.20 |
SFSWAP |
splicing factor, suppressor of white-apricot homolog (Drosophila) |
15831 |
0.2 |
| chr15_52167016_52167539 | 0.20 |
ENSG00000207484 |
. |
9476 |
0.13 |
| chr1_54964656_54964938 | 0.20 |
ENSG00000265404 |
. |
5544 |
0.2 |
| chr1_33207592_33208218 | 0.20 |
KIAA1522 |
KIAA1522 |
419 |
0.8 |
| chr6_155254000_155254239 | 0.20 |
ENSG00000238963 |
. |
27828 |
0.18 |
| chr16_20922512_20922925 | 0.20 |
LYRM1 |
LYR motif containing 1 |
10292 |
0.16 |
| chr5_142206750_142206915 | 0.20 |
ARHGAP26-AS1 |
ARHGAP26 antisense RNA 1 |
41643 |
0.17 |
| chr6_72595878_72596126 | 0.20 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
404 |
0.92 |
| chr16_72281740_72281986 | 0.20 |
PMFBP1 |
polyamine modulated factor 1 binding protein 1 |
71086 |
0.1 |
| chr4_83706873_83707176 | 0.20 |
SCD5 |
stearoyl-CoA desaturase 5 |
12885 |
0.19 |
| chr14_30683326_30683477 | 0.20 |
PRKD1 |
protein kinase D1 |
22297 |
0.28 |
| chr1_201852942_201853093 | 0.20 |
SHISA4 |
shisa family member 4 |
4819 |
0.14 |
| chr5_95294045_95294496 | 0.20 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
3282 |
0.24 |
| chr13_80354703_80354855 | 0.20 |
NDFIP2 |
Nedd4 family interacting protein 2 |
299191 |
0.01 |
| chr1_205568507_205568658 | 0.20 |
ENSG00000206762 |
. |
4413 |
0.18 |
| chr3_16006218_16006514 | 0.20 |
ENSG00000207815 |
. |
91088 |
0.08 |
| chr3_36421358_36421509 | 0.20 |
STAC |
SH3 and cysteine rich domain |
403 |
0.92 |
| chr3_129205233_129205384 | 0.20 |
IFT122 |
intraflagellar transport 122 homolog (Chlamydomonas) |
1726 |
0.3 |
| chr11_48085778_48085988 | 0.19 |
ENSG00000263693 |
. |
32451 |
0.17 |
| chr20_5149549_5150123 | 0.19 |
CDS2 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
9632 |
0.17 |
| chr1_109262925_109263076 | 0.19 |
FNDC7 |
fibronectin type III domain containing 7 |
2033 |
0.27 |
| chr2_136578935_136579086 | 0.19 |
AC011893.3 |
|
1249 |
0.42 |
| chr7_127318408_127318559 | 0.19 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
26249 |
0.16 |
| chr11_48057635_48057786 | 0.19 |
AC103828.1 |
|
20303 |
0.19 |
| chr9_110310779_110311295 | 0.19 |
KLF4 |
Kruppel-like factor 4 (gut) |
58274 |
0.13 |
| chr9_18476779_18476930 | 0.19 |
ADAMTSL1 |
ADAMTS-like 1 |
2623 |
0.41 |
| chr15_88789971_88790162 | 0.19 |
NTRK3 |
neurotrophic tyrosine kinase, receptor, type 3 |
5787 |
0.33 |
| chr15_72529055_72529849 | 0.19 |
PKM |
pyruvate kinase, muscle |
5288 |
0.17 |
| chr13_110971902_110972142 | 0.19 |
COL4A2 |
collagen, type IV, alpha 2 |
12408 |
0.21 |
| chr3_69434805_69435778 | 0.19 |
FRMD4B |
FERM domain containing 4B |
67 |
0.99 |
| chr8_1703891_1704084 | 0.19 |
CTD-2336O2.1 |
|
7476 |
0.21 |
| chr7_100070466_100070929 | 0.19 |
TSC22D4 |
TSC22 domain family, member 4 |
4998 |
0.08 |
| chr10_126845563_126845852 | 0.19 |
CTBP2 |
C-terminal binding protein 2 |
1578 |
0.53 |
| chr9_25677269_25678110 | 0.19 |
TUSC1 |
tumor suppressor candidate 1 |
1167 |
0.68 |
| chr12_54688592_54688942 | 0.19 |
NFE2 |
nuclear factor, erythroid 2 |
775 |
0.4 |
| chr3_134202229_134202939 | 0.19 |
CEP63 |
centrosomal protein 63kDa |
2001 |
0.29 |
| chr3_64211567_64211718 | 0.19 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
511 |
0.83 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.3 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.3 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.2 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.1 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.2 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.3 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0032344 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.2 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.1 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.3 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.3 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0061043 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:1904376 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0031394 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0060346 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.0 | GO:0010658 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.0 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.0 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.0 | 0.1 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.0 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0003283 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.0 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.0 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.1 | GO:0099518 | vesicle transport along microtubule(GO:0047496) vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0090114 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0001738 | morphogenesis of a polarized epithelium(GO:0001738) |
| 0.0 | 0.2 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) negative regulation of lipase activity(GO:0060192) |
| 0.0 | 0.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) negative regulation of cation transmembrane transport(GO:1904063) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.0 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.1 | GO:0006837 | serotonin transport(GO:0006837) |
| 0.0 | 0.0 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.0 | 0.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.0 | GO:0000470 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.4 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.0 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0044309 | neuron spine(GO:0044309) |
| 0.0 | 0.1 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.2 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.0 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.0 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.0 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |