Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
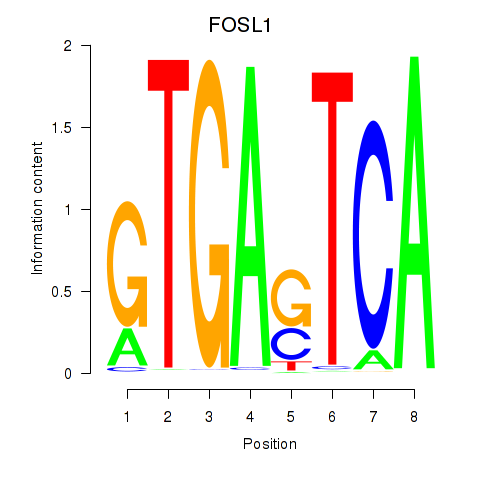
Results for FOSL1
Z-value: 0.40
Transcription factors associated with FOSL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSL1
|
ENSG00000175592.4 | FOS like 1, AP-1 transcription factor subunit |
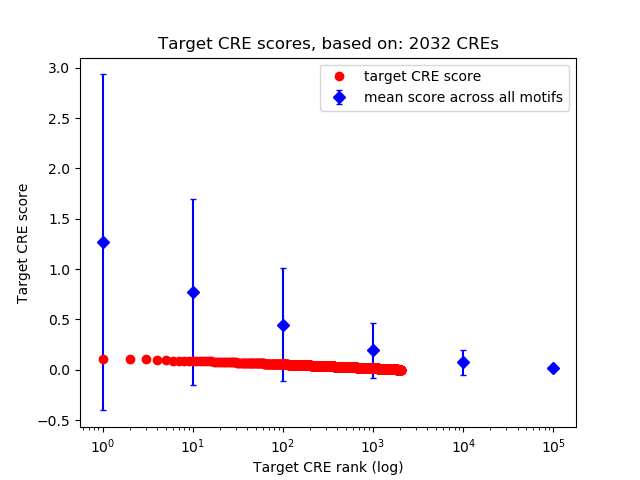
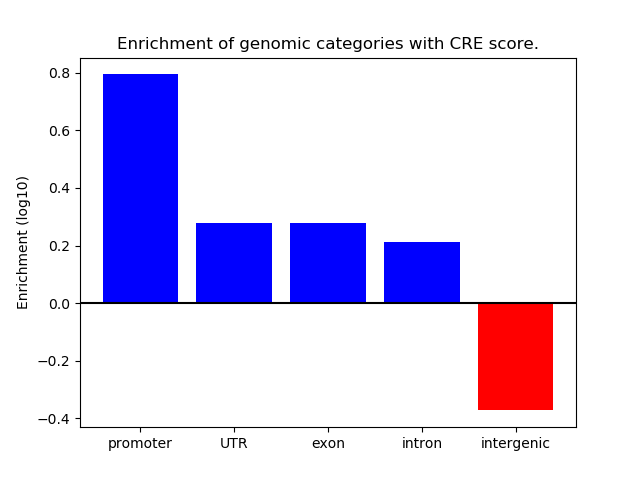
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_65667259_65667446 | FOSL1 | 538 | 0.549011 | 0.58 | 1.0e-01 | Click! |
| chr11_65667492_65667657 | FOSL1 | 316 | 0.748972 | 0.49 | 1.8e-01 | Click! |
| chr11_65667660_65668040 | FOSL1 | 40 | 0.936875 | 0.48 | 1.9e-01 | Click! |
| chr11_65666966_65667198 | FOSL1 | 808 | 0.376641 | 0.36 | 3.4e-01 | Click! |
| chr11_65664383_65664636 | FOSL1 | 3381 | 0.097737 | 0.33 | 3.8e-01 | Click! |
Activity of the FOSL1 motif across conditions
Conditions sorted by the z-value of the FOSL1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
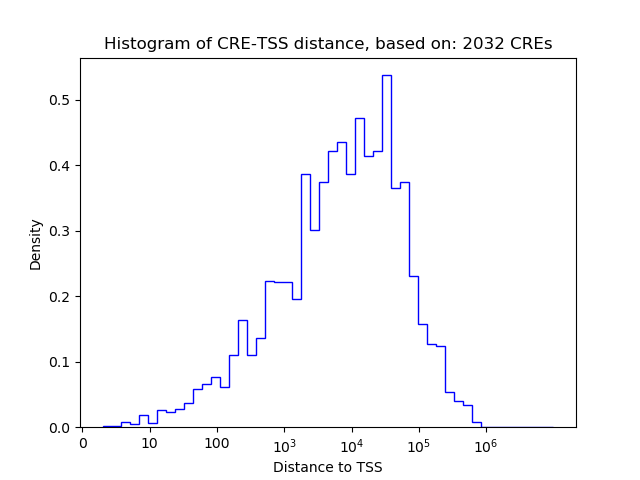
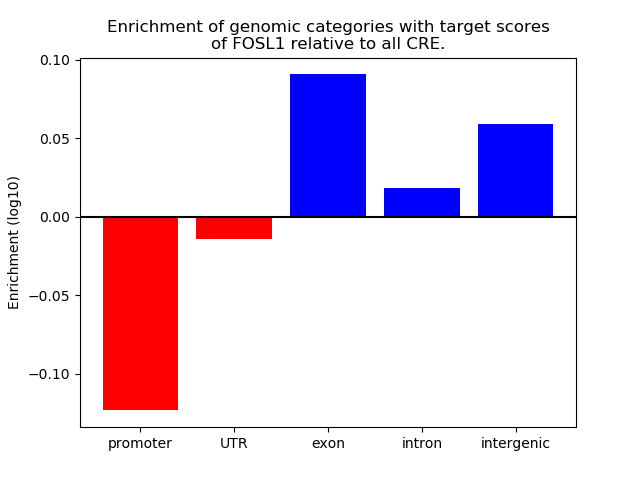
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_21462508_21463481 | 0.11 |
NEBL-AS1 |
NEBL antisense RNA 1 |
51 |
0.59 |
| chr4_40578981_40579556 | 0.10 |
RBM47 |
RNA binding motif protein 47 |
52613 |
0.14 |
| chr17_75439465_75439616 | 0.10 |
ENSG00000200651 |
. |
1349 |
0.37 |
| chr4_15907365_15907907 | 0.10 |
FGFBP1 |
fibroblast growth factor binding protein 1 |
32335 |
0.17 |
| chr2_69059738_69059974 | 0.09 |
AC097495.2 |
|
680 |
0.71 |
| chr3_53303838_53304218 | 0.09 |
ENSG00000238565 |
. |
13291 |
0.12 |
| chr17_75120485_75120909 | 0.09 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
3258 |
0.26 |
| chr16_434070_434221 | 0.09 |
TMEM8A |
transmembrane protein 8A |
2163 |
0.17 |
| chr6_1614377_1614621 | 0.09 |
FOXC1 |
forkhead box C1 |
3818 |
0.36 |
| chr4_185735728_185735879 | 0.09 |
RP11-701P16.2 |
Uncharacterized protein |
1030 |
0.49 |
| chr2_26992134_26992285 | 0.09 |
SLC35F6 |
solute carrier family 35, member F6 |
5052 |
0.2 |
| chr9_71590240_71590555 | 0.09 |
PRKACG |
protein kinase, cAMP-dependent, catalytic, gamma |
38642 |
0.15 |
| chr4_38762729_38763007 | 0.09 |
ENSG00000222230 |
. |
2355 |
0.26 |
| chr1_100865744_100865895 | 0.08 |
ENSG00000216067 |
. |
21488 |
0.18 |
| chr18_9743562_9744068 | 0.08 |
RAB31 |
RAB31, member RAS oncogene family |
35653 |
0.16 |
| chrX_38729814_38730081 | 0.08 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
66811 |
0.12 |
| chr3_169583101_169583252 | 0.08 |
LRRC31 |
leucine rich repeat containing 31 |
4445 |
0.16 |
| chr16_31367909_31368060 | 0.08 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
1441 |
0.29 |
| chr11_2954272_2954423 | 0.08 |
PHLDA2 |
pleckstrin homology-like domain, family A, member 2 |
3662 |
0.15 |
| chr7_143081157_143081800 | 0.08 |
ZYX |
zyxin |
1385 |
0.27 |
| chr19_54603941_54604092 | 0.08 |
OSCAR |
osteoclast associated, immunoglobulin-like receptor |
67 |
0.93 |
| chr8_8945449_8945788 | 0.08 |
ENSG00000239078 |
. |
15592 |
0.15 |
| chr2_111751565_111751824 | 0.08 |
ACOXL |
acyl-CoA oxidase-like |
6919 |
0.31 |
| chr3_46989936_46990087 | 0.08 |
CCDC12 |
coiled-coil domain containing 12 |
28259 |
0.13 |
| chr3_58320901_58321052 | 0.08 |
PXK |
PX domain containing serine/threonine kinase |
1943 |
0.34 |
| chr8_134156595_134156746 | 0.08 |
TG |
thyroglobulin |
30923 |
0.17 |
| chr20_6750709_6750962 | 0.08 |
BMP2 |
bone morphogenetic protein 2 |
2524 |
0.43 |
| chr16_31276706_31276857 | 0.07 |
ENSG00000252876 |
. |
2031 |
0.19 |
| chr19_44179596_44179898 | 0.07 |
PLAUR |
plasminogen activator, urokinase receptor |
5048 |
0.11 |
| chr11_121337447_121337703 | 0.07 |
RP11-730K11.1 |
|
13853 |
0.23 |
| chr10_94520371_94520715 | 0.07 |
ENSG00000201412 |
. |
50178 |
0.13 |
| chr3_149371875_149372212 | 0.07 |
WWTR1-AS1 |
WWTR1 antisense RNA 1 |
2764 |
0.22 |
| chr4_88817282_88817552 | 0.07 |
MEPE |
matrix extracellular phosphoglycoprotein |
63278 |
0.1 |
| chr17_55980324_55980725 | 0.07 |
CUEDC1 |
CUE domain containing 1 |
226 |
0.93 |
| chrX_48956913_48957064 | 0.07 |
WDR45 |
WD repeat domain 45 |
1071 |
0.31 |
| chr6_133006336_133006628 | 0.07 |
VNN1 |
vanin 1 |
28706 |
0.12 |
| chr2_42345389_42345540 | 0.07 |
EML4 |
echinoderm microtubule associated protein like 4 |
51026 |
0.13 |
| chr9_81879361_81879950 | 0.07 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
307033 |
0.01 |
| chr12_54688592_54688942 | 0.07 |
NFE2 |
nuclear factor, erythroid 2 |
775 |
0.4 |
| chr7_28725630_28725928 | 0.07 |
CREB5 |
cAMP responsive element binding protein 5 |
181 |
0.97 |
| chr9_111396808_111397109 | 0.07 |
ACTL7B |
actin-like 7B |
222281 |
0.02 |
| chr11_93270941_93271177 | 0.07 |
SMCO4 |
single-pass membrane protein with coiled-coil domains 4 |
22 |
0.98 |
| chr11_1884799_1884950 | 0.07 |
LSP1 |
lymphocyte-specific protein 1 |
398 |
0.51 |
| chr2_70224516_70224667 | 0.07 |
PCBP1-AS1 |
PCBP1 antisense RNA 1 |
618 |
0.69 |
| chr5_158611818_158612659 | 0.07 |
RNF145 |
ring finger protein 145 |
22404 |
0.16 |
| chr20_48989399_48989550 | 0.07 |
ENSG00000244376 |
. |
56546 |
0.13 |
| chr8_142137516_142137731 | 0.07 |
RP11-809O17.1 |
|
597 |
0.62 |
| chr4_166361643_166362016 | 0.07 |
CPE |
carboxypeptidase E |
22451 |
0.23 |
| chr3_151046365_151046540 | 0.07 |
P2RY13 |
purinergic receptor P2Y, G-protein coupled, 13 |
884 |
0.57 |
| chr1_42420201_42420352 | 0.07 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
35898 |
0.22 |
| chr10_106079197_106079348 | 0.07 |
RP11-127L20.5 |
|
7373 |
0.13 |
| chr6_34283894_34284092 | 0.06 |
C6orf1 |
chromosome 6 open reading frame 1 |
66746 |
0.09 |
| chr8_126308914_126309065 | 0.06 |
ENSG00000242170 |
. |
26143 |
0.21 |
| chr2_37116896_37117561 | 0.06 |
ENSG00000252756 |
. |
22590 |
0.18 |
| chr9_132579196_132579347 | 0.06 |
TOR1A |
torsin family 1, member A (torsin A) |
7142 |
0.14 |
| chr15_58624481_58624632 | 0.06 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
53094 |
0.15 |
| chrX_109083591_109084037 | 0.06 |
ENSG00000252069 |
. |
95836 |
0.08 |
| chr8_23399124_23399403 | 0.06 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
12690 |
0.16 |
| chr4_74573765_74574020 | 0.06 |
IL8 |
interleukin 8 |
32331 |
0.19 |
| chr20_44439937_44440088 | 0.06 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
1203 |
0.27 |
| chr16_89396583_89396806 | 0.06 |
ANKRD11 |
ankyrin repeat domain 11 |
1300 |
0.35 |
| chr11_67917703_67917854 | 0.06 |
CTD-2655K5.1 |
|
20513 |
0.14 |
| chr4_27985809_27985960 | 0.06 |
RP11-180C1.1 |
Uncharacterized protein |
418439 |
0.01 |
| chr17_55945610_55945963 | 0.06 |
CUEDC1 |
CUE domain containing 1 |
1010 |
0.52 |
| chr18_21442082_21442233 | 0.06 |
LAMA3 |
laminin, alpha 3 |
10647 |
0.22 |
| chr2_143828948_143829099 | 0.06 |
ARHGAP15 |
Rho GTPase activating protein 15 |
19908 |
0.24 |
| chr1_161600101_161600252 | 0.06 |
FCGR3B |
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
646 |
0.42 |
| chr19_12899780_12900482 | 0.06 |
JUNB |
jun B proto-oncogene |
2179 |
0.11 |
| chr8_22921887_22922169 | 0.06 |
RP11-875O11.2 |
|
3714 |
0.14 |
| chrX_38728231_38728382 | 0.06 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
65170 |
0.13 |
| chr4_10182968_10183393 | 0.06 |
WDR1 |
WD repeat domain 1 |
64607 |
0.11 |
| chr8_128310206_128310357 | 0.06 |
POU5F1B |
POU class 5 homeobox 1B |
116254 |
0.07 |
| chr1_159061696_159061989 | 0.06 |
AIM2 |
absent in melanoma 2 |
15151 |
0.17 |
| chr9_136037311_136037462 | 0.06 |
GBGT1 |
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 |
1896 |
0.23 |
| chr19_18526600_18526768 | 0.06 |
SSBP4 |
single stranded DNA binding protein 4 |
2990 |
0.12 |
| chr13_31270968_31271119 | 0.06 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
38602 |
0.18 |
| chr4_105982370_105982725 | 0.06 |
ENSG00000252136 |
. |
44597 |
0.16 |
| chr6_24950848_24950999 | 0.06 |
FAM65B |
family with sequence similarity 65, member B |
14735 |
0.22 |
| chr4_79473579_79473973 | 0.06 |
ANXA3 |
annexin A3 |
655 |
0.79 |
| chr4_87932888_87933250 | 0.06 |
AFF1 |
AF4/FMR2 family, member 1 |
4648 |
0.31 |
| chr21_28802590_28802741 | 0.06 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
463833 |
0.01 |
| chr3_101655473_101655624 | 0.06 |
ENSG00000202177 |
. |
18373 |
0.23 |
| chr11_2395489_2395640 | 0.06 |
ENSG00000199550 |
. |
1696 |
0.2 |
| chr15_89710840_89711060 | 0.06 |
RLBP1 |
retinaldehyde binding protein 1 |
44124 |
0.12 |
| chr15_37402275_37402801 | 0.06 |
MEIS2 |
Meis homeobox 2 |
9034 |
0.23 |
| chrX_130199798_130199949 | 0.06 |
ARHGAP36 |
Rho GTPase activating protein 36 |
7089 |
0.23 |
| chr11_85852849_85853000 | 0.06 |
ENSG00000200877 |
. |
11345 |
0.22 |
| chr5_171279662_171279813 | 0.06 |
SMIM23 |
small integral membrane protein 23 |
66847 |
0.11 |
| chr17_27482435_27482735 | 0.06 |
RP11-321A17.4 |
|
15084 |
0.15 |
| chr1_95261518_95261669 | 0.06 |
SLC44A3 |
solute carrier family 44, member 3 |
24305 |
0.19 |
| chr7_127662588_127662739 | 0.06 |
LRRC4 |
leucine rich repeat containing 4 |
8395 |
0.26 |
| chr10_121031057_121031208 | 0.06 |
ENSG00000242853 |
. |
58763 |
0.09 |
| chr1_6659470_6659968 | 0.06 |
KLHL21 |
kelch-like family member 21 |
157 |
0.92 |
| chr1_28502388_28502717 | 0.06 |
PTAFR |
platelet-activating factor receptor |
1147 |
0.35 |
| chr1_44985301_44985452 | 0.06 |
ENSG00000263381 |
. |
25789 |
0.18 |
| chr6_13573909_13574318 | 0.06 |
SIRT5 |
sirtuin 5 |
713 |
0.62 |
| chr9_67026589_67026806 | 0.06 |
ENSG00000265285 |
. |
68236 |
0.13 |
| chr12_57642712_57642903 | 0.06 |
STAC3 |
SH3 and cysteine rich domain 3 |
2160 |
0.17 |
| chr1_151579139_151579290 | 0.05 |
SNX27 |
sorting nexin family member 27 |
5344 |
0.12 |
| chr7_100070466_100070929 | 0.05 |
TSC22D4 |
TSC22 domain family, member 4 |
4998 |
0.08 |
| chr3_50658137_50658523 | 0.05 |
MAPKAPK3 |
mitogen-activated protein kinase-activated protein kinase 3 |
3509 |
0.19 |
| chr13_114830089_114831005 | 0.05 |
RASA3 |
RAS p21 protein activator 3 |
12891 |
0.24 |
| chr1_214649424_214649729 | 0.05 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
11430 |
0.29 |
| chr13_73634006_73635089 | 0.05 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
1617 |
0.45 |
| chr4_166256749_166256900 | 0.05 |
MSMO1 |
methylsterol monooxygenase 1 |
7745 |
0.21 |
| chr12_20522251_20522500 | 0.05 |
PDE3A |
phosphodiesterase 3A, cGMP-inhibited |
196 |
0.89 |
| chr15_99074007_99074158 | 0.05 |
FAM169B |
family with sequence similarity 169, member B |
16471 |
0.27 |
| chr6_35568962_35570321 | 0.05 |
ENSG00000212579 |
. |
49954 |
0.1 |
| chr2_147579520_147579813 | 0.05 |
ENSG00000238860 |
. |
501873 |
0.0 |
| chr15_91459213_91459364 | 0.05 |
MAN2A2 |
mannosidase, alpha, class 2A, member 2 |
3170 |
0.11 |
| chr12_6855415_6855566 | 0.05 |
MLF2 |
myeloid leukemia factor 2 |
6354 |
0.08 |
| chr22_37960000_37960286 | 0.05 |
CDC42EP1 |
CDC42 effector protein (Rho GTPase binding) 1 |
270 |
0.86 |
| chr3_132105351_132105502 | 0.05 |
DNAJC13 |
DnaJ (Hsp40) homolog, subfamily C, member 13 |
30944 |
0.18 |
| chr1_149369357_149369823 | 0.05 |
FCGR1C |
Fc fragment of IgG, high affinity Ic, receptor (CD64), pseudogene |
233 |
0.9 |
| chr22_35697778_35697929 | 0.05 |
TOM1 |
target of myb1 (chicken) |
1966 |
0.25 |
| chr15_51633704_51634417 | 0.05 |
GLDN |
gliomedin |
234 |
0.91 |
| chr1_28189787_28189938 | 0.05 |
THEMIS2 |
thymocyte selection associated family member 2 |
9193 |
0.11 |
| chr1_226830744_226831144 | 0.05 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
16427 |
0.2 |
| chr20_31888809_31889213 | 0.05 |
BPIFB1 |
BPI fold containing family B, member 1 |
18070 |
0.15 |
| chr9_37953977_37954130 | 0.05 |
ENSG00000251745 |
. |
17288 |
0.22 |
| chr9_69655552_69655703 | 0.05 |
AL445665.1 |
Protein LOC100996643 |
5364 |
0.29 |
| chr6_109288373_109288524 | 0.05 |
RP11-787I22.3 |
|
31237 |
0.15 |
| chr10_101908565_101908716 | 0.05 |
ERLIN1 |
ER lipid raft associated 1 |
37146 |
0.11 |
| chr19_12893555_12894002 | 0.05 |
ENSG00000263800 |
. |
4164 |
0.08 |
| chr19_4374737_4374888 | 0.05 |
SH3GL1 |
SH3-domain GRB2-like 1 |
5477 |
0.08 |
| chr2_218559547_218559837 | 0.05 |
DIRC3 |
disrupted in renal carcinoma 3 |
61586 |
0.14 |
| chr9_42853652_42853803 | 0.05 |
AC129778.2 |
|
2565 |
0.39 |
| chr3_194875945_194876317 | 0.05 |
XXYLT1-AS2 |
XXYLT1 antisense RNA 2 |
7511 |
0.15 |
| chr3_12504067_12504243 | 0.05 |
TSEN2 |
TSEN2 tRNA splicing endonuclease subunit |
21776 |
0.15 |
| chr20_48909337_48909488 | 0.05 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
102036 |
0.06 |
| chr1_176180985_176181136 | 0.05 |
RP11-195C7.1 |
|
4276 |
0.23 |
| chr22_24903141_24903292 | 0.05 |
AP000355.2 |
|
9260 |
0.13 |
| chr4_166143196_166143347 | 0.05 |
KLHL2 |
kelch-like family member 2 |
2118 |
0.32 |
| chr2_64440045_64440196 | 0.05 |
AC074289.1 |
|
6011 |
0.28 |
| chr11_46697090_46697391 | 0.05 |
ATG13 |
autophagy related 13 |
11690 |
0.13 |
| chr18_57569513_57570014 | 0.05 |
PMAIP1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
2515 |
0.37 |
| chr11_119583713_119583864 | 0.05 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
15475 |
0.18 |
| chr1_33807573_33808014 | 0.05 |
ENSG00000222112 |
. |
5328 |
0.13 |
| chr19_1168749_1168988 | 0.05 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
74 |
0.95 |
| chr16_85096756_85096907 | 0.05 |
KIAA0513 |
KIAA0513 |
13 |
0.98 |
| chr14_70059974_70060446 | 0.05 |
KIAA0247 |
KIAA0247 |
18103 |
0.2 |
| chr5_96126813_96126964 | 0.05 |
CTD-2260A17.1 |
|
6224 |
0.15 |
| chr8_129262963_129263114 | 0.05 |
ENSG00000201782 |
. |
30288 |
0.23 |
| chr3_112930660_112930893 | 0.05 |
BOC |
BOC cell adhesion associated, oncogene regulated |
266 |
0.93 |
| chr4_108745530_108746004 | 0.05 |
SGMS2 |
sphingomyelin synthase 2 |
48 |
0.98 |
| chrX_9881450_9881696 | 0.05 |
SHROOM2 |
shroom family member 2 |
850 |
0.65 |
| chr5_7676594_7676745 | 0.05 |
ADCY2 |
adenylate cyclase 2 (brain) |
22605 |
0.24 |
| chr3_15310922_15311111 | 0.05 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
6400 |
0.15 |
| chr3_42138838_42138989 | 0.05 |
TRAK1 |
trafficking protein, kinesin binding 1 |
6351 |
0.29 |
| chr9_114614096_114614247 | 0.05 |
ENSG00000206907 |
. |
12063 |
0.2 |
| chr1_36816210_36816695 | 0.05 |
STK40 |
serine/threonine kinase 40 |
10489 |
0.13 |
| chr5_100113349_100113842 | 0.05 |
ENSG00000221263 |
. |
38674 |
0.19 |
| chr6_18488896_18489181 | 0.05 |
ENSG00000207775 |
. |
82977 |
0.09 |
| chr7_29153330_29153633 | 0.05 |
CPVL |
carboxypeptidase, vitellogenic-like |
97 |
0.97 |
| chr1_109373299_109373450 | 0.05 |
AKNAD1 |
AKNA domain containing 1 |
21965 |
0.15 |
| chr19_11449251_11449402 | 0.05 |
RAB3D |
RAB3D, member RAS oncogene family |
1018 |
0.31 |
| chr9_132251498_132251652 | 0.05 |
ENSG00000264298 |
. |
10740 |
0.21 |
| chr19_39889302_39889453 | 0.05 |
MED29 |
mediator complex subunit 29 |
7325 |
0.08 |
| chr4_141077504_141077678 | 0.05 |
MAML3 |
mastermind-like 3 (Drosophila) |
2253 |
0.36 |
| chr16_279128_279418 | 0.05 |
LUC7L |
LUC7-like (S. cerevisiae) |
148 |
0.91 |
| chr7_20824481_20824853 | 0.05 |
SP8 |
Sp8 transcription factor |
1838 |
0.51 |
| chr8_11706405_11706556 | 0.05 |
RP11-589N15.2 |
|
2580 |
0.22 |
| chr9_6417233_6417502 | 0.05 |
UHRF2 |
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase |
3664 |
0.25 |
| chr19_11202980_11203131 | 0.05 |
LDLR |
low density lipoprotein receptor |
1780 |
0.29 |
| chr4_1716841_1717066 | 0.05 |
SLBP |
stem-loop binding protein |
2671 |
0.19 |
| chr7_102990167_102990648 | 0.05 |
PSMC2 |
proteasome (prosome, macropain) 26S subunit, ATPase, 2 |
2297 |
0.25 |
| chr15_92619299_92619450 | 0.05 |
RP11-24J19.1 |
|
96292 |
0.09 |
| chr6_42301055_42301206 | 0.05 |
ENSG00000221252 |
. |
79004 |
0.08 |
| chr10_114746275_114746635 | 0.05 |
RP11-57H14.2 |
|
34821 |
0.18 |
| chr1_23912147_23912298 | 0.05 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
25937 |
0.15 |
| chr7_36603678_36603829 | 0.05 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
30509 |
0.17 |
| chr2_139274196_139274347 | 0.05 |
SPOPL |
speckle-type POZ protein-like |
14900 |
0.31 |
| chr5_6705329_6705480 | 0.05 |
PAPD7 |
PAP associated domain containing 7 |
9314 |
0.26 |
| chr6_14744303_14744454 | 0.05 |
ENSG00000206960 |
. |
97612 |
0.09 |
| chr2_28177663_28177868 | 0.05 |
ENSG00000265321 |
. |
41469 |
0.15 |
| chr12_54585165_54585441 | 0.04 |
SMUG1 |
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
2525 |
0.18 |
| chrX_87167581_87167854 | 0.04 |
ENSG00000252016 |
. |
236076 |
0.02 |
| chr12_3277666_3277860 | 0.04 |
TSPAN9-IT1 |
TSPAN9 intronic transcript 1 (non-protein coding) |
18800 |
0.21 |
| chr10_35927790_35928179 | 0.04 |
ENSG00000264780 |
. |
2196 |
0.26 |
| chr10_45872200_45872351 | 0.04 |
ALOX5 |
arachidonate 5-lipoxygenase |
2600 |
0.34 |
| chr1_149754304_149754780 | 0.04 |
FCGR1A |
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
296 |
0.78 |
| chr14_75590762_75591162 | 0.04 |
NEK9 |
NIMA-related kinase 9 |
2781 |
0.19 |
| chr2_173557887_173558038 | 0.04 |
ENSG00000239041 |
. |
1935 |
0.35 |
| chr7_55620313_55620510 | 0.04 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
134 |
0.97 |
| chr19_2169086_2169237 | 0.04 |
DOT1L |
DOT1-like histone H3K79 methyltransferase |
4977 |
0.13 |
| chr2_191278529_191278680 | 0.04 |
MFSD6 |
major facilitator superfamily domain containing 6 |
5523 |
0.22 |
| chr22_30642991_30643200 | 0.04 |
LIF |
leukemia inhibitory factor |
255 |
0.78 |
| chr19_49071767_49072143 | 0.04 |
SULT2B1 |
sulfotransferase family, cytosolic, 2B, member 1 |
6842 |
0.11 |
| chr11_64099501_64099713 | 0.04 |
AP003774.1 |
|
2631 |
0.11 |
| chr9_67293693_67293844 | 0.04 |
RP11-236F9.2 |
|
8470 |
0.29 |
| chr8_142141679_142142152 | 0.04 |
RP11-809O17.1 |
|
1855 |
0.33 |
| chrX_109124093_109124244 | 0.04 |
TMEM164 |
transmembrane protein 164 |
121695 |
0.05 |
| chr9_124044853_124045004 | 0.04 |
RP11-477J21.6 |
|
80 |
0.96 |
| chr15_43531850_43532099 | 0.04 |
ENSG00000202211 |
. |
7904 |
0.13 |
| chr8_38856565_38856716 | 0.04 |
ADAM9 |
ADAM metallopeptidase domain 9 |
2135 |
0.22 |
| chr19_47853304_47853535 | 0.04 |
DHX34 |
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
881 |
0.52 |
| chrX_147691631_147691782 | 0.04 |
AFF2-IT1 |
AFF2 intronic transcript 1 (non-protein coding) |
63307 |
0.14 |
| chr15_80452796_80453143 | 0.04 |
FAH |
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
4676 |
0.25 |
| chr11_65667660_65668040 | 0.04 |
FOSL1 |
FOS-like antigen 1 |
40 |
0.94 |
| chr22_40832446_40832730 | 0.04 |
RP5-1042K10.13 |
|
21581 |
0.14 |
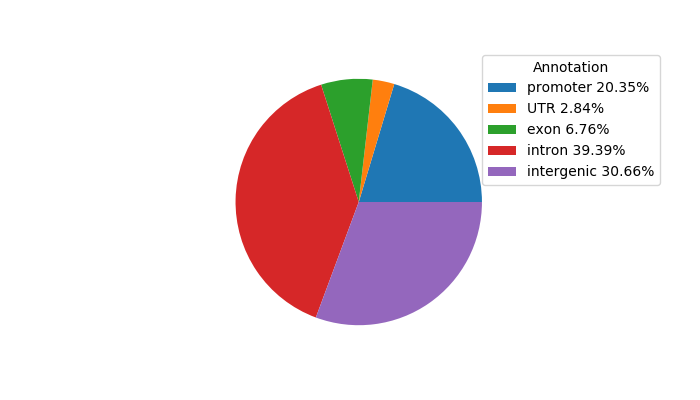
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0051459 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.0 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |