Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
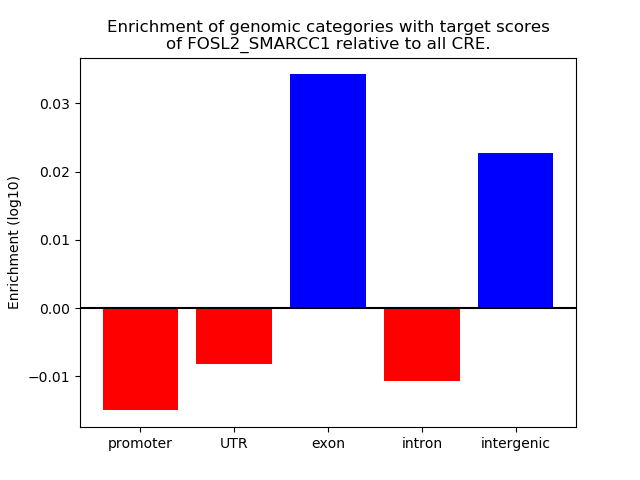
Results for FOSL2_SMARCC1
Z-value: 1.04
Transcription factors associated with FOSL2_SMARCC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
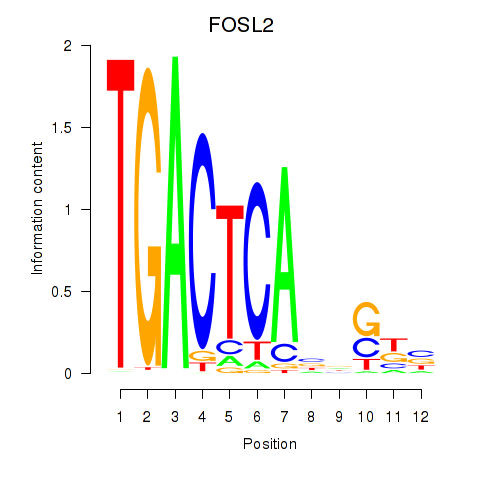
FOSL2
|
ENSG00000075426.7 | FOS like 2, AP-1 transcription factor subunit |
|
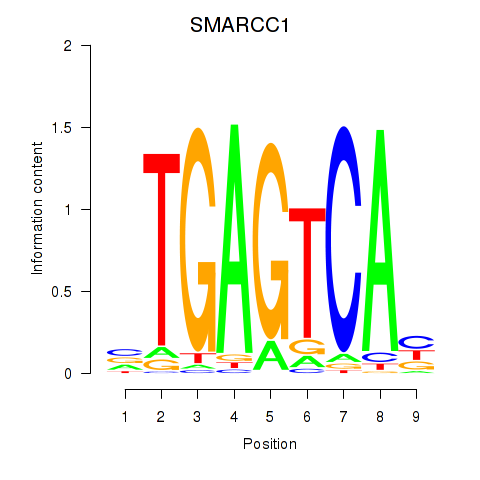
SMARCC1
|
ENSG00000173473.6 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_28589849_28590028 | FOSL2 | 25731 | 0.150297 | 0.88 | 1.6e-03 | Click! |
| chr2_28582365_28582516 | FOSL2 | 33229 | 0.138157 | 0.87 | 2.5e-03 | Click! |
| chr2_28601116_28601352 | FOSL2 | 14435 | 0.165249 | 0.81 | 8.2e-03 | Click! |
| chr2_28582657_28582808 | FOSL2 | 32937 | 0.138655 | 0.79 | 1.1e-02 | Click! |
| chr2_28600514_28600665 | FOSL2 | 15080 | 0.164501 | 0.75 | 2.0e-02 | Click! |
| chr3_47822404_47822881 | SMARCC1 | 765 | 0.620272 | 0.60 | 8.5e-02 | Click! |
| chr3_47823051_47823226 | SMARCC1 | 269 | 0.903870 | 0.50 | 1.7e-01 | Click! |
| chr3_47831109_47831260 | SMARCC1 | 7777 | 0.168002 | -0.34 | 3.7e-01 | Click! |
| chr3_47823573_47824113 | SMARCC1 | 436 | 0.811272 | 0.33 | 3.9e-01 | Click! |
| chr3_47822128_47822402 | SMARCC1 | 1142 | 0.460753 | 0.29 | 4.4e-01 | Click! |
Activity of the FOSL2_SMARCC1 motif across conditions
Conditions sorted by the z-value of the FOSL2_SMARCC1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
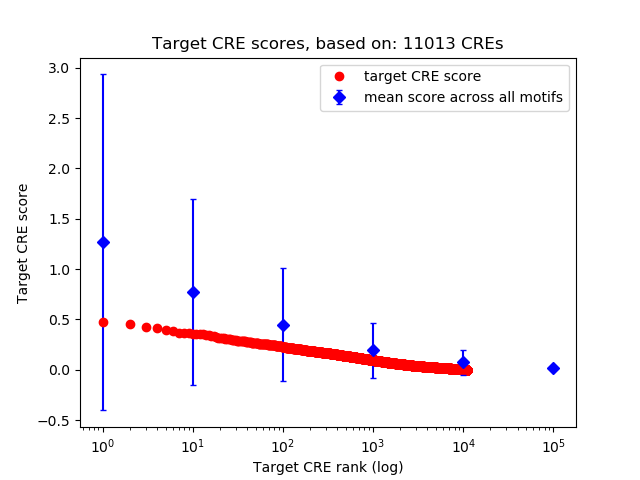
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_150436563_150436714 | 0.48 |
TNIP1 |
TNFAIP3 interacting protein 1 |
8018 |
0.2 |
| chr8_8945449_8945788 | 0.46 |
ENSG00000239078 |
. |
15592 |
0.15 |
| chr18_12005840_12005991 | 0.43 |
RP11-820I16.3 |
|
10978 |
0.16 |
| chr19_4374737_4374888 | 0.42 |
SH3GL1 |
SH3-domain GRB2-like 1 |
5477 |
0.08 |
| chr1_2266428_2266579 | 0.40 |
AL589739.1 |
Uncharacterized protein |
7922 |
0.11 |
| chr14_103801113_103802028 | 0.39 |
EIF5 |
eukaryotic translation initiation factor 5 |
284 |
0.87 |
| chr3_27410282_27410923 | 0.37 |
NEK10 |
NIMA-related kinase 10 |
281 |
0.91 |
| chr6_35453862_35454013 | 0.36 |
TEAD3 |
TEA domain family member 3 |
6038 |
0.17 |
| chr10_121031057_121031208 | 0.36 |
ENSG00000242853 |
. |
58763 |
0.09 |
| chr2_202015949_202016344 | 0.36 |
CFLAR-AS1 |
CFLAR antisense RNA 1 |
263 |
0.88 |
| chr17_73567230_73567381 | 0.36 |
MYO15B |
myosin XVB pseudogene |
19553 |
0.1 |
| chr2_95940298_95940449 | 0.35 |
PROM2 |
prominin 2 |
128 |
0.97 |
| chr19_2089959_2090185 | 0.35 |
MOB3A |
MOB kinase activator 3A |
103 |
0.94 |
| chr7_100781654_100781965 | 0.35 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
11430 |
0.1 |
| chr4_10182968_10183393 | 0.35 |
WDR1 |
WD repeat domain 1 |
64607 |
0.11 |
| chr11_64099501_64099713 | 0.34 |
AP003774.1 |
|
2631 |
0.11 |
| chr13_114830089_114831005 | 0.33 |
RASA3 |
RAS p21 protein activator 3 |
12891 |
0.24 |
| chr7_36603678_36603829 | 0.33 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
30509 |
0.17 |
| chr17_74495503_74495654 | 0.32 |
RHBDF2 |
rhomboid 5 homolog 2 (Drosophila) |
1875 |
0.2 |
| chr15_91459213_91459364 | 0.32 |
MAN2A2 |
mannosidase, alpha, class 2A, member 2 |
3170 |
0.11 |
| chr6_35568962_35570321 | 0.32 |
ENSG00000212579 |
. |
49954 |
0.1 |
| chr16_31145770_31145921 | 0.31 |
PRSS8 |
protease, serine, 8 |
1116 |
0.22 |
| chr2_231191950_231192650 | 0.31 |
SP140L |
SP140 nuclear body protein-like |
315 |
0.92 |
| chr17_79798852_79799080 | 0.31 |
RP11-498C9.2 |
|
2075 |
0.12 |
| chr1_30856346_30856497 | 0.30 |
MATN1-AS1 |
MATN1 antisense RNA 1 |
334930 |
0.01 |
| chr10_45870440_45870591 | 0.30 |
ALOX5 |
arachidonate 5-lipoxygenase |
840 |
0.69 |
| chr1_198610920_198611071 | 0.30 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
2703 |
0.34 |
| chr19_12897015_12897166 | 0.30 |
ENSG00000263800 |
. |
852 |
0.31 |
| chr2_147579520_147579813 | 0.30 |
ENSG00000238860 |
. |
501873 |
0.0 |
| chr17_4396255_4396406 | 0.30 |
SPNS2 |
spinster homolog 2 (Drosophila) |
5803 |
0.13 |
| chr11_11589938_11590089 | 0.29 |
RP11-483L5.1 |
|
1618 |
0.47 |
| chr8_96237876_96238027 | 0.29 |
C8orf37 |
chromosome 8 open reading frame 37 |
43478 |
0.17 |
| chr4_170001005_170001626 | 0.29 |
RP11-483A20.3 |
|
69734 |
0.1 |
| chr12_109084399_109085395 | 0.29 |
CORO1C |
coronin, actin binding protein, 1C |
5281 |
0.17 |
| chr1_151579139_151579290 | 0.29 |
SNX27 |
sorting nexin family member 27 |
5344 |
0.12 |
| chr9_138022409_138022560 | 0.28 |
OLFM1 |
olfactomedin 1 |
34652 |
0.19 |
| chr17_8976590_8976741 | 0.28 |
NTN1 |
netrin 1 |
51045 |
0.15 |
| chr1_155102127_155102278 | 0.28 |
EFNA1 |
ephrin-A1 |
1850 |
0.14 |
| chr17_2869143_2869294 | 0.28 |
CTD-3060P21.1 |
|
29 |
0.98 |
| chr4_15907365_15907907 | 0.28 |
FGFBP1 |
fibroblast growth factor binding protein 1 |
32335 |
0.17 |
| chr19_39894109_39894559 | 0.28 |
ZFP36 |
ZFP36 ring finger protein |
3119 |
0.1 |
| chr5_74964815_74965486 | 0.28 |
ENSG00000207333 |
. |
40274 |
0.14 |
| chr12_25207808_25208034 | 0.28 |
LRMP |
lymphoid-restricted membrane protein |
2247 |
0.32 |
| chr20_61455814_61456027 | 0.27 |
COL9A3 |
collagen, type IX, alpha 3 |
6757 |
0.12 |
| chr16_88366626_88367008 | 0.27 |
ZNF469 |
zinc finger protein 469 |
127062 |
0.05 |
| chr2_99069202_99069353 | 0.27 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
7864 |
0.24 |
| chrX_47489439_47489603 | 0.27 |
CFP |
complement factor properdin |
157 |
0.92 |
| chr3_186993064_186993215 | 0.27 |
MASP1 |
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
16346 |
0.21 |
| chr11_104914891_104915295 | 0.26 |
CARD16 |
caspase recruitment domain family, member 16 |
941 |
0.54 |
| chrX_38729814_38730081 | 0.26 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
66811 |
0.12 |
| chr4_79473579_79473973 | 0.26 |
ANXA3 |
annexin A3 |
655 |
0.79 |
| chr19_48246517_48246668 | 0.26 |
GLTSCR2 |
glioma tumor suppressor candidate region gene 2 |
2187 |
0.21 |
| chr20_52519916_52520067 | 0.26 |
AC005220.3 |
|
36708 |
0.2 |
| chr8_10512332_10512483 | 0.26 |
RP1L1 |
retinitis pigmentosa 1-like 1 |
205 |
0.92 |
| chr14_89345487_89345638 | 0.26 |
TTC8 |
tetratricopeptide repeat domain 8 |
37690 |
0.15 |
| chr3_126206096_126206282 | 0.26 |
ZXDC |
ZXD family zinc finger C |
11427 |
0.16 |
| chr11_11592510_11592661 | 0.26 |
RP11-483L5.1 |
|
954 |
0.65 |
| chr17_80175660_80175811 | 0.26 |
RP13-516M14.2 |
|
3632 |
0.13 |
| chrX_133147131_133147282 | 0.26 |
GPC3 |
glypican 3 |
27284 |
0.21 |
| chr3_187491731_187491915 | 0.26 |
BCL6 |
B-cell CLL/lymphoma 6 |
28308 |
0.2 |
| chr22_32058342_32058690 | 0.26 |
PISD |
phosphatidylserine decarboxylase |
98 |
0.97 |
| chr16_31276706_31276857 | 0.26 |
ENSG00000252876 |
. |
2031 |
0.19 |
| chr1_25361354_25361505 | 0.26 |
ENSG00000264371 |
. |
11435 |
0.23 |
| chr1_244486436_244486726 | 0.25 |
C1orf100 |
chromosome 1 open reading frame 100 |
29356 |
0.21 |
| chr9_132579196_132579347 | 0.25 |
TOR1A |
torsin family 1, member A (torsin A) |
7142 |
0.14 |
| chr1_52159244_52159395 | 0.25 |
OSBPL9 |
oxysterol binding protein-like 9 |
24096 |
0.16 |
| chr16_54459980_54460131 | 0.25 |
ENSG00000264079 |
. |
123253 |
0.06 |
| chr11_2395489_2395640 | 0.25 |
ENSG00000199550 |
. |
1696 |
0.2 |
| chr1_156630677_156631393 | 0.25 |
RP11-284F21.7 |
|
181 |
0.89 |
| chr11_46378662_46378813 | 0.25 |
DGKZ |
diacylglycerol kinase, zeta |
4408 |
0.16 |
| chr21_28802590_28802741 | 0.25 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
463833 |
0.01 |
| chr11_129697989_129698558 | 0.25 |
TMEM45B |
transmembrane protein 45B |
12559 |
0.26 |
| chr7_64407394_64407545 | 0.25 |
ZNF273 |
zinc finger protein 273 |
43835 |
0.11 |
| chr1_248069833_248069987 | 0.25 |
OR2W3 |
olfactory receptor, family 2, subfamily W, member 3 |
11021 |
0.1 |
| chr14_96312809_96312960 | 0.25 |
ENSG00000263357 |
. |
5043 |
0.29 |
| chr21_31012601_31012752 | 0.25 |
RP11-313P22.1 |
|
17050 |
0.22 |
| chr17_6757690_6757943 | 0.24 |
TEKT1 |
tektin 1 |
22736 |
0.14 |
| chrX_105065104_105065255 | 0.24 |
NRK |
Nik related kinase |
1357 |
0.55 |
| chr1_41363272_41363423 | 0.24 |
RP5-1066H13.4 |
|
33761 |
0.14 |
| chr2_109237700_109238010 | 0.24 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
133 |
0.97 |
| chr11_10695962_10696877 | 0.24 |
MRVI1 |
murine retrovirus integration site 1 homolog |
18642 |
0.17 |
| chr4_105982370_105982725 | 0.24 |
ENSG00000252136 |
. |
44597 |
0.16 |
| chr22_46409167_46409603 | 0.24 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
36376 |
0.09 |
| chr10_45872200_45872351 | 0.24 |
ALOX5 |
arachidonate 5-lipoxygenase |
2600 |
0.34 |
| chr15_89630929_89631312 | 0.24 |
RP11-326A19.3 |
|
52 |
0.77 |
| chr4_27985809_27985960 | 0.24 |
RP11-180C1.1 |
Uncharacterized protein |
418439 |
0.01 |
| chr4_74922619_74922770 | 0.24 |
PPBPP2 |
pro-platelet basic protein pseudogene 2 |
2157 |
0.24 |
| chr11_1884799_1884950 | 0.24 |
LSP1 |
lymphocyte-specific protein 1 |
398 |
0.51 |
| chr14_75085047_75085411 | 0.24 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
5923 |
0.21 |
| chr3_46989936_46990087 | 0.23 |
CCDC12 |
coiled-coil domain containing 12 |
28259 |
0.13 |
| chr6_147197427_147197865 | 0.23 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
25505 |
0.25 |
| chr14_35871775_35873043 | 0.23 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
937 |
0.68 |
| chr1_30602704_30603087 | 0.23 |
ENSG00000222787 |
. |
245146 |
0.02 |
| chr17_78865183_78865610 | 0.23 |
RPTOR |
regulatory associated protein of MTOR, complex 1 |
31128 |
0.13 |
| chr11_2198846_2198997 | 0.23 |
ENSG00000265258 |
. |
4628 |
0.12 |
| chr1_160983030_160983181 | 0.23 |
F11R |
F11 receptor |
7781 |
0.11 |
| chr17_4762471_4762622 | 0.23 |
ENSG00000263599 |
. |
16375 |
0.08 |
| chr10_112086583_112086734 | 0.23 |
SMNDC1 |
survival motor neuron domain containing 1 |
21949 |
0.22 |
| chr14_92997841_92998087 | 0.23 |
RIN3 |
Ras and Rab interactor 3 |
17816 |
0.26 |
| chr19_48784148_48784299 | 0.23 |
ZNF114 |
zinc finger protein 114 |
9343 |
0.11 |
| chr17_75439465_75439616 | 0.23 |
ENSG00000200651 |
. |
1349 |
0.37 |
| chr5_176874674_176875204 | 0.23 |
PRR7-AS1 |
PRR7 antisense RNA 1 |
239 |
0.62 |
| chr12_111847113_111847630 | 0.23 |
SH2B3 |
SH2B adaptor protein 3 |
3619 |
0.23 |
| chr2_219725294_219725445 | 0.23 |
WNT6 |
wingless-type MMTV integration site family, member 6 |
825 |
0.5 |
| chr17_75138230_75138411 | 0.22 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
852 |
0.63 |
| chr1_235093242_235093393 | 0.22 |
ENSG00000239690 |
. |
53384 |
0.15 |
| chr17_39779992_39780143 | 0.22 |
KRT17 |
keratin 17 |
547 |
0.62 |
| chr22_43288275_43288426 | 0.22 |
ARFGAP3 |
ADP-ribosylation factor GTPase activating protein 3 |
34238 |
0.17 |
| chr2_25966238_25966389 | 0.22 |
ENSG00000222070 |
. |
21252 |
0.18 |
| chr3_93878024_93878443 | 0.22 |
ENSG00000253062 |
. |
60823 |
0.12 |
| chr3_64963602_64963753 | 0.22 |
ENSG00000264871 |
. |
254981 |
0.02 |
| chr10_45870253_45870404 | 0.22 |
ALOX5 |
arachidonate 5-lipoxygenase |
653 |
0.77 |
| chr17_55945610_55945963 | 0.22 |
CUEDC1 |
CUE domain containing 1 |
1010 |
0.52 |
| chr3_53303838_53304218 | 0.22 |
ENSG00000238565 |
. |
13291 |
0.12 |
| chr9_111396808_111397109 | 0.22 |
ACTL7B |
actin-like 7B |
222281 |
0.02 |
| chr20_1470626_1470777 | 0.22 |
SIRPB2 |
signal-regulatory protein beta 2 |
1341 |
0.29 |
| chr10_54463336_54463751 | 0.22 |
RP11-556E13.1 |
|
51626 |
0.16 |
| chr8_68515024_68515175 | 0.22 |
ENSG00000221660 |
. |
108768 |
0.07 |
| chr1_10961126_10961370 | 0.22 |
C1orf127 |
chromosome 1 open reading frame 127 |
46679 |
0.12 |
| chr8_108509260_108510202 | 0.22 |
ANGPT1 |
angiopoietin 1 |
507 |
0.89 |
| chr19_35951714_35951865 | 0.22 |
FFAR2 |
free fatty acid receptor 2 |
11172 |
0.11 |
| chr6_35338140_35338402 | 0.22 |
PPARD |
peroxisome proliferator-activated receptor delta |
27869 |
0.16 |
| chr2_111751565_111751824 | 0.22 |
ACOXL |
acyl-CoA oxidase-like |
6919 |
0.31 |
| chr8_128310206_128310357 | 0.21 |
POU5F1B |
POU class 5 homeobox 1B |
116254 |
0.07 |
| chr20_16800308_16800459 | 0.21 |
OTOR |
otoraplin |
71380 |
0.11 |
| chr19_36248323_36248705 | 0.21 |
HSPB6 |
heat shock protein, alpha-crystallin-related, B6 |
466 |
0.38 |
| chr3_31247061_31247212 | 0.21 |
ENSG00000222983 |
. |
23071 |
0.23 |
| chr7_20824481_20824853 | 0.21 |
SP8 |
Sp8 transcription factor |
1838 |
0.51 |
| chr8_11706405_11706556 | 0.21 |
RP11-589N15.2 |
|
2580 |
0.22 |
| chr7_155090835_155091229 | 0.21 |
INSIG1 |
insulin induced gene 1 |
761 |
0.7 |
| chr16_70780347_70780498 | 0.21 |
RP11-394B2.6 |
|
538 |
0.68 |
| chr16_11753228_11753379 | 0.21 |
SNN |
stannin |
8967 |
0.18 |
| chr12_121363997_121364148 | 0.21 |
SPPL3 |
signal peptide peptidase like 3 |
21898 |
0.14 |
| chr3_5055062_5055354 | 0.21 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
33562 |
0.15 |
| chr7_1553073_1553492 | 0.21 |
AC102953.6 |
|
4219 |
0.16 |
| chr6_133006336_133006628 | 0.21 |
VNN1 |
vanin 1 |
28706 |
0.12 |
| chr13_34116508_34116659 | 0.21 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
191816 |
0.03 |
| chr1_36808540_36808801 | 0.21 |
STK40 |
serine/threonine kinase 40 |
18271 |
0.12 |
| chr9_130717791_130717942 | 0.21 |
FAM102A |
family with sequence similarity 102, member A |
4865 |
0.11 |
| chr6_18488896_18489181 | 0.21 |
ENSG00000207775 |
. |
82977 |
0.09 |
| chr20_49983111_49983262 | 0.21 |
ENSG00000263645 |
. |
10672 |
0.28 |
| chr5_10632325_10632658 | 0.21 |
ANKRD33B-AS1 |
ANKRD33B antisense RNA 1 |
4154 |
0.28 |
| chr2_206633742_206633893 | 0.21 |
AC007362.3 |
|
5087 |
0.3 |
| chr13_50114703_50115207 | 0.21 |
RCBTB1 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
26504 |
0.17 |
| chr14_50468177_50468632 | 0.21 |
C14orf182 |
chromosome 14 open reading frame 182 |
5834 |
0.2 |
| chrX_19757821_19757972 | 0.21 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
1059 |
0.68 |
| chr10_95143500_95143651 | 0.20 |
MYOF |
myoferlin |
98376 |
0.07 |
| chr14_70120086_70120237 | 0.20 |
KIAA0247 |
KIAA0247 |
41848 |
0.16 |
| chr2_134415347_134415498 | 0.20 |
ENSG00000200708 |
. |
61760 |
0.14 |
| chr2_8655367_8655518 | 0.20 |
AC011747.7 |
|
160454 |
0.04 |
| chr15_89710840_89711060 | 0.20 |
RLBP1 |
retinaldehyde binding protein 1 |
44124 |
0.12 |
| chr5_14298302_14298453 | 0.20 |
TRIO |
trio Rho guanine nucleotide exchange factor |
7291 |
0.33 |
| chr6_160041713_160041864 | 0.20 |
SOD2 |
superoxide dismutase 2, mitochondrial |
72472 |
0.09 |
| chr21_46310753_46310904 | 0.20 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
17076 |
0.1 |
| chr7_33080072_33080440 | 0.20 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
265 |
0.9 |
| chr11_9385409_9385962 | 0.20 |
IPO7 |
importin 7 |
20484 |
0.16 |
| chr1_6086993_6087271 | 0.20 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
225 |
0.93 |
| chr13_76867689_76867887 | 0.20 |
ENSG00000243274 |
. |
158721 |
0.04 |
| chr6_139870784_139870935 | 0.20 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
175102 |
0.03 |
| chr11_35188283_35188434 | 0.20 |
CD44 |
CD44 molecule (Indian blood group) |
9760 |
0.17 |
| chr5_118609514_118610204 | 0.20 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
5410 |
0.21 |
| chr22_32033546_32033968 | 0.20 |
PISD |
phosphatidylserine decarboxylase |
634 |
0.74 |
| chr14_35092634_35092920 | 0.20 |
SNX6 |
sorting nexin 6 |
6101 |
0.2 |
| chr18_3602703_3603732 | 0.20 |
DLGAP1-AS2 |
DLGAP1 antisense RNA 2 |
219 |
0.92 |
| chr16_89429989_89430201 | 0.20 |
ANKRD11 |
ankyrin repeat domain 11 |
34701 |
0.1 |
| chr7_20255346_20255644 | 0.20 |
MACC1 |
metastasis associated in colon cancer 1 |
1518 |
0.49 |
| chr10_8099206_8099457 | 0.20 |
GATA3 |
GATA binding protein 3 |
2562 |
0.44 |
| chr9_94186787_94187440 | 0.20 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
969 |
0.69 |
| chr15_27495001_27495152 | 0.20 |
GABRG3 |
gamma-aminobutyric acid (GABA) A receptor, gamma 3 |
21613 |
0.2 |
| chr11_35061934_35062494 | 0.20 |
PDHX |
pyruvate dehydrogenase complex, component X |
62883 |
0.1 |
| chr1_162667094_162667245 | 0.20 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
64909 |
0.1 |
| chr7_73119920_73120071 | 0.20 |
ENSG00000265724 |
. |
5652 |
0.11 |
| chrX_149352977_149353128 | 0.20 |
ENSG00000252454 |
. |
43187 |
0.19 |
| chr1_51069078_51069376 | 0.20 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
8943 |
0.23 |
| chr4_166361643_166362016 | 0.20 |
CPE |
carboxypeptidase E |
22451 |
0.23 |
| chr9_128441842_128441993 | 0.20 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
6995 |
0.28 |
| chrX_48917537_48917688 | 0.19 |
CCDC120 |
coiled-coil domain containing 120 |
1066 |
0.3 |
| chr9_132648263_132648472 | 0.19 |
FNBP1 |
formin binding protein 1 |
33222 |
0.12 |
| chr1_9399387_9399538 | 0.19 |
SPSB1 |
splA/ryanodine receptor domain and SOCS box containing 1 |
12028 |
0.22 |
| chr5_158611818_158612659 | 0.19 |
RNF145 |
ring finger protein 145 |
22404 |
0.16 |
| chr19_42388528_42389094 | 0.19 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
296 |
0.84 |
| chr14_33883349_33883500 | 0.19 |
NPAS3 |
neuronal PAS domain protein 3 |
198906 |
0.03 |
| chr12_3277666_3277860 | 0.19 |
TSPAN9-IT1 |
TSPAN9 intronic transcript 1 (non-protein coding) |
18800 |
0.21 |
| chr10_134330839_134330990 | 0.19 |
LINC01165 |
long intergenic non-protein coding RNA 1165 |
5039 |
0.22 |
| chrX_40294011_40294465 | 0.19 |
ENSG00000206677 |
. |
57854 |
0.14 |
| chr19_34311073_34311611 | 0.19 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
13513 |
0.27 |
| chr6_34071312_34071463 | 0.19 |
GRM4 |
glutamate receptor, metabotropic 4 |
244 |
0.94 |
| chr16_88837085_88837352 | 0.19 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
14401 |
0.08 |
| chr22_46929020_46929201 | 0.19 |
CELSR1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
2081 |
0.33 |
| chr1_6659470_6659968 | 0.19 |
KLHL21 |
kelch-like family member 21 |
157 |
0.92 |
| chr9_127023581_127024115 | 0.19 |
NEK6 |
NIMA-related kinase 6 |
96 |
0.82 |
| chr9_132251498_132251652 | 0.19 |
ENSG00000264298 |
. |
10740 |
0.21 |
| chr3_122486679_122486830 | 0.19 |
ENSG00000238480 |
. |
17535 |
0.17 |
| chr19_2169086_2169237 | 0.19 |
DOT1L |
DOT1-like histone H3K79 methyltransferase |
4977 |
0.13 |
| chr17_56410108_56410593 | 0.19 |
MIR142 |
microRNA 142 |
481 |
0.66 |
| chr8_128911768_128911919 | 0.19 |
TMEM75 |
transmembrane protein 75 |
48748 |
0.14 |
| chr7_33078848_33079779 | 0.19 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
1208 |
0.42 |
| chr19_39889302_39889453 | 0.19 |
MED29 |
mediator complex subunit 29 |
7325 |
0.08 |
| chr2_111875565_111875871 | 0.19 |
BCL2L11 |
BCL2-like 11 (apoptosis facilitator) |
1237 |
0.5 |
| chr2_70224516_70224667 | 0.19 |
PCBP1-AS1 |
PCBP1 antisense RNA 1 |
618 |
0.69 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.2 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.1 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.1 | 0.3 | GO:0071501 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 0.2 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.1 | 0.3 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.2 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.0 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.4 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.2 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0048343 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.3 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.3 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.0 | 0.0 | GO:0043096 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.0 | GO:0014742 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.1 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 0.0 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0002885 | positive regulation of hypersensitivity(GO:0002885) |
| 0.0 | 0.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:1901374 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.0 | 0.1 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.0 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:1902931 | negative regulation of alcohol biosynthetic process(GO:1902931) |
| 0.0 | 0.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.0 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0009304 | tRNA transcription(GO:0009304) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.2 | GO:0009309 | amine biosynthetic process(GO:0009309) |
| 0.0 | 0.3 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 0.1 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.0 | GO:0051136 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.0 | GO:0046015 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:2001235 | positive regulation of mitochondrion organization(GO:0010822) positive regulation of release of cytochrome c from mitochondria(GO:0090200) positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0032753 | positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 | 0.0 | GO:0097502 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.0 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0071331 | cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.1 | GO:0043558 | regulation of translational initiation in response to stress(GO:0043558) |
| 0.0 | 0.0 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.0 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.0 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.0 | GO:0033185 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.0 | GO:0036126 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |