Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
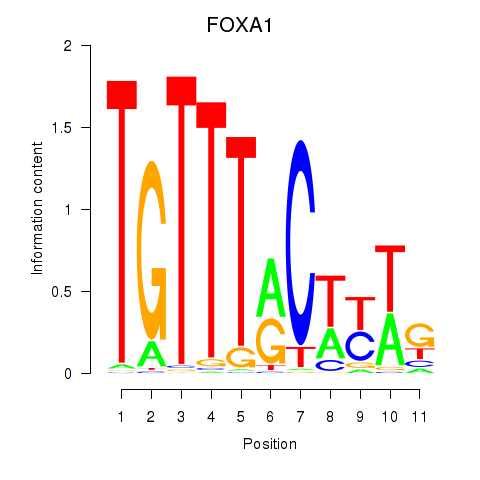
Results for FOXA1
Z-value: 1.51
Transcription factors associated with FOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA1
|
ENSG00000129514.4 | forkhead box A1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_38016039_38016231 | FOXA1 | 48104 | 0.149801 | 0.71 | 3.3e-02 | Click! |
| chr14_38063381_38063532 | FOXA1 | 783 | 0.608797 | 0.65 | 5.9e-02 | Click! |
| chr14_38052851_38053204 | FOXA1 | 11212 | 0.225216 | 0.59 | 9.5e-02 | Click! |
| chr14_38063567_38064244 | FOXA1 | 334 | 0.835480 | 0.57 | 1.1e-01 | Click! |
| chr14_38068776_38069062 | FOXA1 | 326 | 0.905613 | 0.50 | 1.7e-01 | Click! |
Activity of the FOXA1 motif across conditions
Conditions sorted by the z-value of the FOXA1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
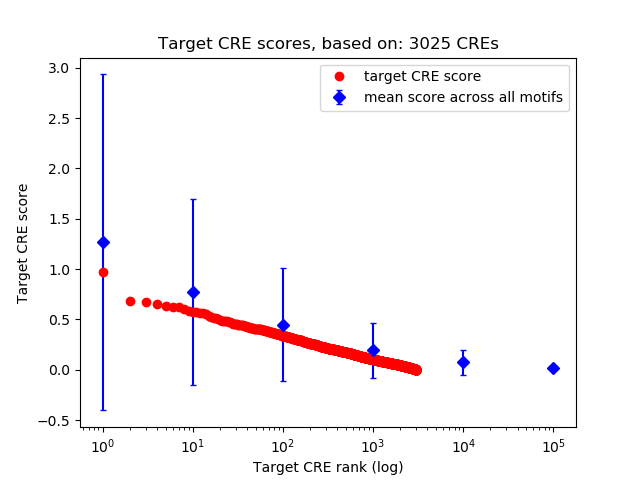
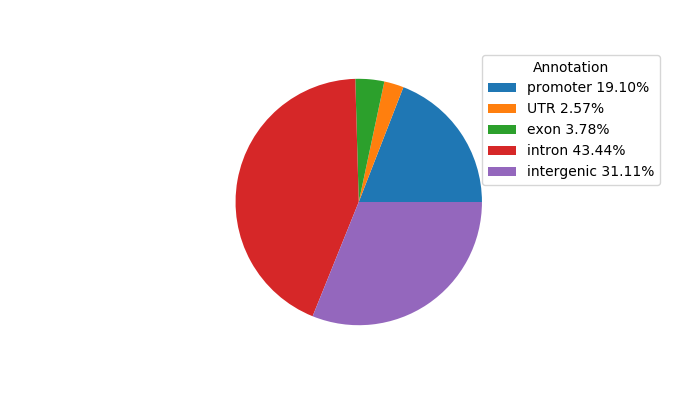
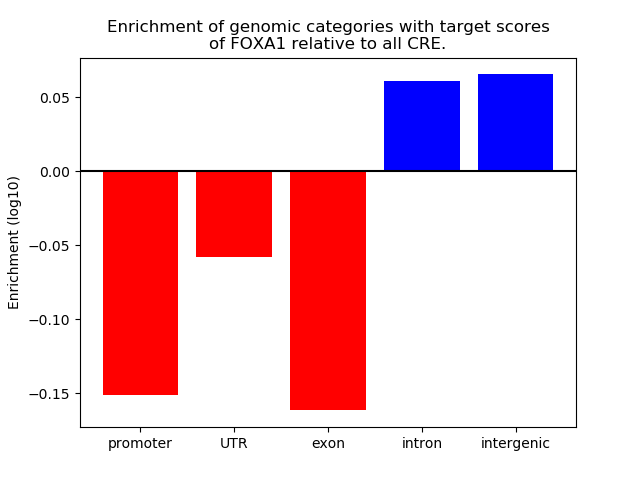
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr21_28214648_28215240 | 0.97 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
393 |
0.9 |
| chr7_102470506_102470794 | 0.68 |
ENSG00000252643 |
. |
4402 |
0.21 |
| chr4_48701412_48701950 | 0.68 |
FRYL |
FRY-like |
18493 |
0.24 |
| chr5_72746721_72747193 | 0.65 |
FOXD1 |
forkhead box D1 |
2605 |
0.29 |
| chr15_96878987_96879612 | 0.63 |
ENSG00000222651 |
. |
2809 |
0.21 |
| chr1_61668583_61669016 | 0.63 |
RP4-802A10.1 |
|
78394 |
0.11 |
| chr4_81392876_81393298 | 0.62 |
C4orf22 |
chromosome 4 open reading frame 22 |
136164 |
0.05 |
| chr2_151334778_151334948 | 0.60 |
RND3 |
Rho family GTPase 3 |
7033 |
0.34 |
| chr3_124604600_124604823 | 0.58 |
ITGB5 |
integrin, beta 5 |
1433 |
0.47 |
| chr1_215179577_215179737 | 0.58 |
KCNK2 |
potassium channel, subfamily K, member 2 |
459 |
0.91 |
| chr2_230035602_230035855 | 0.57 |
PID1 |
phosphotyrosine interaction domain containing 1 |
61073 |
0.14 |
| chr8_125438541_125438825 | 0.56 |
TRMT12 |
tRNA methyltransferase 12 homolog (S. cerevisiae) |
24365 |
0.16 |
| chr2_46165643_46166035 | 0.56 |
PRKCE |
protein kinase C, epsilon |
62202 |
0.14 |
| chr2_150418995_150419256 | 0.55 |
AC144449.1 |
|
24585 |
0.19 |
| chr2_190647319_190647470 | 0.53 |
ORMDL1 |
ORM1-like 1 (S. cerevisiae) |
714 |
0.49 |
| chr6_17418741_17419244 | 0.52 |
CAP2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
25049 |
0.25 |
| chr12_59478573_59478971 | 0.52 |
RP11-150C16.1 |
|
164352 |
0.04 |
| chr7_27217405_27217623 | 0.51 |
RP1-170O19.20 |
Uncharacterized protein |
2118 |
0.1 |
| chr2_38265076_38265999 | 0.51 |
RMDN2-AS1 |
RMDN2 antisense RNA 1 |
2053 |
0.33 |
| chr12_56920749_56920947 | 0.50 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
5065 |
0.15 |
| chr6_10426220_10426432 | 0.49 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
6455 |
0.18 |
| chr6_53457738_53458227 | 0.48 |
GCLC |
glutamate-cysteine ligase, catalytic subunit |
23786 |
0.17 |
| chr3_100429715_100429877 | 0.48 |
TFG |
TRK-fused gene |
982 |
0.64 |
| chr4_123795073_123795451 | 0.48 |
RP11-170N16.3 |
|
10605 |
0.18 |
| chr15_33585949_33586454 | 0.48 |
RP11-489D6.2 |
|
16246 |
0.22 |
| chr6_151648337_151648657 | 0.47 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
1674 |
0.27 |
| chr10_25155972_25156509 | 0.46 |
ENSG00000240294 |
. |
41501 |
0.18 |
| chr1_94701701_94701935 | 0.46 |
ARHGAP29 |
Rho GTPase activating protein 29 |
1303 |
0.53 |
| chr1_82267832_82267983 | 0.46 |
LPHN2 |
latrophilin 2 |
1825 |
0.52 |
| chr9_113799034_113800104 | 0.46 |
LPAR1 |
lysophosphatidic acid receptor 1 |
754 |
0.74 |
| chr2_133998643_133998815 | 0.45 |
AC010890.1 |
|
23811 |
0.24 |
| chr1_65363518_65363760 | 0.45 |
JAK1 |
Janus kinase 1 |
68548 |
0.12 |
| chr3_149865487_149865870 | 0.45 |
RP11-167H9.4 |
|
49859 |
0.15 |
| chr7_94025115_94025296 | 0.44 |
COL1A2 |
collagen, type I, alpha 2 |
1332 |
0.57 |
| chr1_168231073_168231252 | 0.44 |
ENSG00000206880 |
. |
1557 |
0.39 |
| chr6_3825964_3826270 | 0.44 |
RP11-420L9.4 |
|
6050 |
0.22 |
| chr2_151336714_151337073 | 0.43 |
RND3 |
Rho family GTPase 3 |
5003 |
0.36 |
| chr5_103680813_103681290 | 0.43 |
ENSG00000239808 |
. |
84524 |
0.11 |
| chr22_36841557_36841862 | 0.43 |
ENSG00000252225 |
. |
4694 |
0.17 |
| chr12_48395931_48396147 | 0.42 |
COL2A1 |
collagen, type II, alpha 1 |
2065 |
0.24 |
| chr15_99864758_99865035 | 0.42 |
AC022819.2 |
Uncharacterized protein |
5068 |
0.23 |
| chr6_138427325_138427625 | 0.42 |
PERP |
PERP, TP53 apoptosis effector |
1173 |
0.59 |
| chr18_52763849_52764148 | 0.42 |
ENSG00000252437 |
. |
49878 |
0.16 |
| chr1_158087708_158088406 | 0.42 |
KIRREL |
kin of IRRE like (Drosophila) |
31587 |
0.18 |
| chr4_176922498_176923243 | 0.41 |
GPM6A |
glycoprotein M6A |
613 |
0.78 |
| chr6_125698814_125699135 | 0.41 |
RP11-735G4.1 |
|
3504 |
0.35 |
| chr7_41735925_41736162 | 0.41 |
INHBA-AS1 |
INHBA antisense RNA 1 |
2497 |
0.29 |
| chr21_45139141_45140358 | 0.41 |
PDXK |
pyridoxal (pyridoxine, vitamin B6) kinase |
756 |
0.66 |
| chr18_72509166_72509431 | 0.41 |
ZNF407 |
zinc finger protein 407 |
166322 |
0.04 |
| chr15_96869491_96869741 | 0.41 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
449 |
0.57 |
| chr4_187857953_187858132 | 0.41 |
ENSG00000252382 |
. |
79432 |
0.12 |
| chr8_42063953_42065062 | 0.41 |
PLAT |
plasminogen activator, tissue |
576 |
0.71 |
| chr4_124339787_124340099 | 0.41 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
18820 |
0.3 |
| chr3_167811230_167811432 | 0.40 |
GOLIM4 |
golgi integral membrane protein 4 |
1801 |
0.51 |
| chr3_161632471_161632704 | 0.40 |
OTOL1 |
otolin 1 |
417991 |
0.01 |
| chr11_121593168_121593875 | 0.40 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
132393 |
0.05 |
| chr2_150186573_150186784 | 0.40 |
LYPD6 |
LY6/PLAUR domain containing 6 |
179 |
0.97 |
| chr12_115174256_115174747 | 0.40 |
ENSG00000252459 |
. |
1008 |
0.64 |
| chr18_57332899_57333095 | 0.40 |
CCBE1 |
collagen and calcium binding EGF domains 1 |
397 |
0.86 |
| chr5_162992519_162993506 | 0.40 |
MAT2B |
methionine adenosyltransferase II, beta |
60392 |
0.11 |
| chr6_104981791_104982069 | 0.39 |
ENSG00000252944 |
. |
232767 |
0.02 |
| chr6_121758735_121759079 | 0.39 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
2069 |
0.31 |
| chr1_216773992_216774229 | 0.39 |
ESRRG |
estrogen-related receptor gamma |
122564 |
0.06 |
| chr1_185703938_185704134 | 0.39 |
HMCN1 |
hemicentin 1 |
353 |
0.92 |
| chrX_10737482_10738236 | 0.39 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
92080 |
0.1 |
| chr2_66723532_66723851 | 0.39 |
MEIS1 |
Meis homeobox 1 |
12368 |
0.22 |
| chr7_79081974_79082229 | 0.39 |
ENSG00000234456 |
. |
97 |
0.92 |
| chr3_114477226_114477377 | 0.38 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
486 |
0.86 |
| chr4_145570819_145570970 | 0.38 |
HHIP |
hedgehog interacting protein |
3570 |
0.28 |
| chr10_13931045_13931263 | 0.38 |
FRMD4A |
FERM domain containing 4A |
30242 |
0.2 |
| chr21_32591500_32591929 | 0.38 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
57369 |
0.16 |
| chr4_187648403_187648661 | 0.38 |
FAT1 |
FAT atypical cadherin 1 |
656 |
0.82 |
| chr9_33506853_33507187 | 0.38 |
RP11-255A11.2 |
|
7751 |
0.17 |
| chr1_92317111_92317363 | 0.38 |
TGFBR3 |
transforming growth factor, beta receptor III |
9913 |
0.22 |
| chr5_140801311_140801579 | 0.37 |
PCDHGA11 |
protocadherin gamma subfamily A, 11 |
650 |
0.41 |
| chr7_97952929_97953353 | 0.37 |
RP11-307C18.1 |
|
976 |
0.55 |
| chr5_91888200_91888432 | 0.37 |
ENSG00000264489 |
. |
146572 |
0.05 |
| chr10_29824157_29824642 | 0.37 |
ENSG00000207612 |
. |
9627 |
0.19 |
| chr6_45523679_45523878 | 0.36 |
ENSG00000252738 |
. |
90063 |
0.09 |
| chr3_47155829_47156131 | 0.36 |
ENSG00000251938 |
. |
25882 |
0.16 |
| chr2_216296824_216297216 | 0.36 |
FN1 |
fibronectin 1 |
3770 |
0.26 |
| chr15_100583304_100583570 | 0.36 |
ENSG00000252957 |
. |
30958 |
0.16 |
| chr11_16023597_16024065 | 0.36 |
CTD-3096P4.1 |
|
20905 |
0.28 |
| chr6_113994010_113994191 | 0.36 |
ENSG00000221559 |
. |
31935 |
0.2 |
| chr1_172154318_172154604 | 0.36 |
DNM3OS |
DNM3 opposite strand/antisense RNA |
40527 |
0.14 |
| chr3_145879077_145879748 | 0.36 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
458 |
0.86 |
| chr4_157868586_157868737 | 0.36 |
PDGFC |
platelet derived growth factor C |
23394 |
0.2 |
| chr4_184369530_184369754 | 0.35 |
CDKN2AIP |
CDKN2A interacting protein |
3792 |
0.23 |
| chr4_169491131_169491310 | 0.35 |
PALLD |
palladin, cytoskeletal associated protein |
58501 |
0.12 |
| chr8_39939130_39939407 | 0.35 |
C8orf4 |
chromosome 8 open reading frame 4 |
71721 |
0.11 |
| chr4_75175100_75175487 | 0.35 |
EPGN |
epithelial mitogen |
479 |
0.82 |
| chr16_72325870_72326093 | 0.35 |
ENSG00000207514 |
. |
84622 |
0.08 |
| chr1_101754265_101754416 | 0.35 |
RP4-575N6.5 |
|
45626 |
0.13 |
| chr3_16218692_16218887 | 0.35 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
2573 |
0.36 |
| chr9_111034250_111034449 | 0.34 |
ENSG00000222512 |
. |
86860 |
0.1 |
| chr1_205270236_205270504 | 0.34 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
20513 |
0.15 |
| chr22_40706246_40706565 | 0.34 |
ADSL |
adenylosuccinate lyase |
36102 |
0.14 |
| chr13_36049085_36049236 | 0.34 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
1672 |
0.32 |
| chr7_30637504_30637655 | 0.34 |
ENSG00000196295 |
. |
3154 |
0.23 |
| chr13_73712912_73713063 | 0.34 |
ENSG00000265959 |
. |
14040 |
0.19 |
| chr6_157274340_157274740 | 0.34 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
52033 |
0.17 |
| chr12_66089013_66089164 | 0.33 |
HMGA2 |
high mobility group AT-hook 2 |
128823 |
0.05 |
| chr3_124605343_124605563 | 0.33 |
ITGB5 |
integrin, beta 5 |
691 |
0.74 |
| chr18_74237053_74237772 | 0.33 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
3200 |
0.24 |
| chr1_86045810_86046121 | 0.33 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
479 |
0.77 |
| chr6_128828666_128828900 | 0.33 |
RP1-86D1.4 |
|
2513 |
0.24 |
| chr12_15038397_15038786 | 0.33 |
MGP |
matrix Gla protein |
200 |
0.92 |
| chr12_16762141_16762326 | 0.33 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
573 |
0.83 |
| chr13_103382043_103382374 | 0.33 |
CCDC168 |
coiled-coil domain containing 168 |
6951 |
0.13 |
| chr4_41217997_41218148 | 0.33 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
1437 |
0.44 |
| chr5_151062319_151062545 | 0.33 |
SPARC |
secreted protein, acidic, cysteine-rich (osteonectin) |
2359 |
0.22 |
| chr6_82755621_82755792 | 0.33 |
ENSG00000223044 |
. |
164451 |
0.04 |
| chr8_49468858_49469448 | 0.33 |
RP11-770E5.1 |
|
5026 |
0.35 |
| chr12_76653631_76654077 | 0.33 |
ENSG00000223273 |
. |
55677 |
0.14 |
| chr7_73671230_73671468 | 0.32 |
ENSG00000252538 |
. |
1597 |
0.31 |
| chr5_73110322_73110752 | 0.32 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
1194 |
0.51 |
| chr12_13350846_13351208 | 0.32 |
EMP1 |
epithelial membrane protein 1 |
1307 |
0.55 |
| chr13_76278728_76279091 | 0.32 |
LMO7 |
LIM domain 7 |
55594 |
0.13 |
| chr12_13025379_13025989 | 0.32 |
GPRC5A |
G protein-coupled receptor, family C, group 5, member A |
18032 |
0.14 |
| chr6_157639179_157639343 | 0.32 |
ENSG00000252609 |
. |
73170 |
0.11 |
| chr8_48571824_48572102 | 0.32 |
SPIDR |
scaffolding protein involved in DNA repair |
263 |
0.93 |
| chr1_24621056_24621364 | 0.32 |
ENSG00000266511 |
. |
14865 |
0.15 |
| chr2_20643812_20644209 | 0.31 |
RHOB |
ras homolog family member B |
2825 |
0.27 |
| chr19_31714740_31714922 | 0.31 |
AC020952.1 |
Uncharacterized protein |
74469 |
0.12 |
| chr3_98613191_98613556 | 0.31 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
6642 |
0.19 |
| chr2_145188780_145188931 | 0.31 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
718 |
0.79 |
| chr7_121170130_121170295 | 0.31 |
ENSG00000221690 |
. |
44596 |
0.19 |
| chr6_143995099_143995343 | 0.31 |
PHACTR2 |
phosphatase and actin regulator 2 |
3881 |
0.32 |
| chr1_86078649_86078931 | 0.31 |
ENSG00000199934 |
. |
20827 |
0.18 |
| chr1_107684305_107684780 | 0.31 |
NTNG1 |
netrin G1 |
881 |
0.74 |
| chr1_214397865_214398571 | 0.31 |
SMYD2 |
SET and MYND domain containing 2 |
56358 |
0.16 |
| chr5_92917804_92917955 | 0.31 |
ENSG00000237187 |
. |
876 |
0.48 |
| chr1_158969050_158969253 | 0.31 |
IFI16 |
interferon, gamma-inducible protein 16 |
607 |
0.76 |
| chr20_22384501_22385014 | 0.31 |
FOXA2 |
forkhead box A2 |
180344 |
0.03 |
| chr3_109884338_109884502 | 0.31 |
ENSG00000221206 |
. |
386392 |
0.01 |
| chr13_101202971_101203237 | 0.31 |
RP11-151A6.4 |
|
11044 |
0.19 |
| chr5_112398331_112398555 | 0.30 |
DCP2 |
decapping mRNA 2 |
85964 |
0.08 |
| chr17_7515414_7515712 | 0.30 |
ENSG00000251860 |
. |
1064 |
0.18 |
| chr4_174102838_174102989 | 0.30 |
RP11-10K16.1 |
|
12110 |
0.19 |
| chr6_52217103_52217563 | 0.30 |
PAQR8 |
progestin and adipoQ receptor family member VIII |
8886 |
0.23 |
| chr11_91714959_91715138 | 0.30 |
FAT3 |
FAT atypical cadherin 3 |
370214 |
0.01 |
| chr2_106260790_106261225 | 0.30 |
NCK2 |
NCK adaptor protein 2 |
100347 |
0.08 |
| chr2_205411353_205411504 | 0.30 |
PARD3B |
par-3 family cell polarity regulator beta |
705 |
0.83 |
| chr11_92029031_92029279 | 0.30 |
FAT3 |
FAT atypical cadherin 3 |
56107 |
0.17 |
| chr5_132385315_132385509 | 0.30 |
HSPA4 |
heat shock 70kDa protein 4 |
2242 |
0.29 |
| chr9_14423961_14424182 | 0.30 |
NFIB |
nuclear factor I/B |
25089 |
0.25 |
| chr15_63178723_63178874 | 0.30 |
RP11-1069G10.1 |
|
2565 |
0.34 |
| chr12_42978890_42979099 | 0.30 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
4484 |
0.31 |
| chr10_24752381_24752532 | 0.30 |
KIAA1217 |
KIAA1217 |
3004 |
0.32 |
| chr12_19591912_19592176 | 0.30 |
AEBP2 |
AE binding protein 2 |
564 |
0.8 |
| chr12_50608566_50608717 | 0.30 |
RP3-405J10.4 |
|
4746 |
0.13 |
| chr17_2308923_2309089 | 0.29 |
MNT |
MAX network transcriptional repressor |
4594 |
0.13 |
| chr2_1734119_1734469 | 0.29 |
PXDN |
peroxidasin homolog (Drosophila) |
13920 |
0.26 |
| chr3_152046405_152046556 | 0.29 |
TMEM14E |
transmembrane protein 14E |
12299 |
0.22 |
| chr13_36048764_36048946 | 0.29 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
1977 |
0.3 |
| chr1_203159153_203159304 | 0.29 |
CHI3L1 |
chitinase 3-like 1 (cartilage glycoprotein-39) |
3351 |
0.18 |
| chr2_36924555_36924939 | 0.29 |
VIT |
vitrin |
727 |
0.75 |
| chr17_8976227_8976400 | 0.29 |
NTN1 |
netrin 1 |
50693 |
0.15 |
| chr17_45982661_45983102 | 0.29 |
SP2 |
Sp2 transcription factor |
9365 |
0.1 |
| chr3_197021883_197022040 | 0.29 |
ENSG00000265850 |
. |
1142 |
0.4 |
| chr4_2960219_2960405 | 0.29 |
NOP14 |
NOP14 nucleolar protein |
4748 |
0.16 |
| chr6_148762563_148762871 | 0.29 |
ENSG00000223322 |
. |
82659 |
0.1 |
| chr9_110253387_110253828 | 0.29 |
KLF4 |
Kruppel-like factor 4 (gut) |
844 |
0.66 |
| chr8_29027576_29027748 | 0.29 |
ENSG00000264328 |
. |
27452 |
0.15 |
| chr4_182568542_182568740 | 0.28 |
ENSG00000251742 |
. |
187620 |
0.03 |
| chr14_50787669_50787839 | 0.28 |
ATP5S |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
1802 |
0.29 |
| chr5_3600517_3600892 | 0.28 |
CTD-2012M11.3 |
|
402 |
0.89 |
| chr16_87098321_87098755 | 0.28 |
RP11-899L11.3 |
|
150983 |
0.04 |
| chr3_16217443_16217716 | 0.28 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
1363 |
0.53 |
| chr4_169770371_169770556 | 0.28 |
RP11-635L1.3 |
|
16375 |
0.18 |
| chr20_1103594_1103745 | 0.28 |
PSMF1 |
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
4389 |
0.25 |
| chr6_141884450_141884959 | 0.28 |
ENSG00000222764 |
. |
77155 |
0.13 |
| chr14_29228686_29229081 | 0.28 |
RP11-966I7.1 |
|
5586 |
0.17 |
| chr2_102383869_102384043 | 0.28 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
29770 |
0.24 |
| chr4_87503159_87503484 | 0.28 |
MAPK10 |
mitogen-activated protein kinase 10 |
11942 |
0.21 |
| chr5_131893791_131893942 | 0.28 |
RAD50 |
RAD50 homolog (S. cerevisiae) |
943 |
0.42 |
| chr22_43166170_43166377 | 0.28 |
ENSG00000200448 |
. |
7308 |
0.21 |
| chr7_2842191_2842738 | 0.28 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
12428 |
0.25 |
| chr21_17442999_17443232 | 0.28 |
ENSG00000252273 |
. |
35286 |
0.24 |
| chr4_77067485_77067636 | 0.27 |
NUP54 |
nucleoporin 54kDa |
1990 |
0.3 |
| chr3_29388286_29388437 | 0.27 |
ENSG00000216169 |
. |
22551 |
0.22 |
| chr1_81703498_81704139 | 0.27 |
ENSG00000223026 |
. |
13656 |
0.28 |
| chr2_180293372_180293844 | 0.27 |
ZNF385B |
zinc finger protein 385B |
133707 |
0.05 |
| chr5_65097013_65097416 | 0.27 |
NLN |
neurolysin (metallopeptidase M3 family) |
13024 |
0.24 |
| chr6_4059667_4059856 | 0.27 |
ENSG00000252668 |
. |
16862 |
0.13 |
| chr9_97951408_97952029 | 0.27 |
ENSG00000238746 |
. |
16838 |
0.19 |
| chr4_134071254_134071890 | 0.27 |
PCDH10 |
protocadherin 10 |
1102 |
0.7 |
| chr2_174495860_174496019 | 0.27 |
CDCA7 |
cell division cycle associated 7 |
276339 |
0.01 |
| chr12_132316032_132316383 | 0.27 |
MMP17 |
matrix metallopeptidase 17 (membrane-inserted) |
3122 |
0.22 |
| chr3_11267791_11267952 | 0.27 |
HRH1 |
histamine receptor H1 |
154 |
0.97 |
| chr2_63416100_63416251 | 0.27 |
OTX1 |
orthodenticle homeobox 1 |
138238 |
0.05 |
| chr5_111016644_111016835 | 0.27 |
ENSG00000253057 |
. |
39019 |
0.16 |
| chr20_60163995_60164193 | 0.27 |
CDH4 |
cadherin 4, type 1, R-cadherin (retinal) |
10740 |
0.3 |
| chr13_110957687_110957838 | 0.27 |
COL4A2 |
collagen, type IV, alpha 2 |
397 |
0.84 |
| chr4_124981640_124982053 | 0.27 |
ANKRD50 |
ankyrin repeat domain 50 |
652041 |
0.0 |
| chr18_73167828_73168275 | 0.26 |
SMIM21 |
small integral membrane protein 21 |
28393 |
0.22 |
| chr1_218337432_218337583 | 0.26 |
ENSG00000201493 |
. |
34370 |
0.22 |
| chr8_35235141_35235292 | 0.26 |
UNC5D |
unc-5 homolog D (C. elegans) |
141923 |
0.05 |
| chr2_199480864_199481015 | 0.26 |
ENSG00000252511 |
. |
49486 |
0.2 |
| chr15_96886136_96886425 | 0.26 |
ENSG00000222651 |
. |
9790 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.2 | 0.6 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.2 | 0.2 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.2 | 1.1 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.4 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.2 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.3 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.2 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.1 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.2 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.1 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.2 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.3 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.2 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0072071 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.1 | GO:0003171 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.3 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.3 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.2 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:2000696 | nephron tubule epithelial cell differentiation(GO:0072160) regulation of nephron tubule epithelial cell differentiation(GO:0072182) regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0071801 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.2 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0006533 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.0 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.2 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.0 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.0 | GO:0072189 | positive regulation of smooth muscle cell differentiation(GO:0051152) ureter development(GO:0072189) ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.0 | GO:0001738 | morphogenesis of a polarized epithelium(GO:0001738) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.2 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.3 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.0 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0031272 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.2 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0003170 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.0 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.0 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.0 | 0.0 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.4 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.0 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0098644 | collagen type VI trimer(GO:0005589) complex of collagen trimers(GO:0098644) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.0 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.7 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.1 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.2 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |