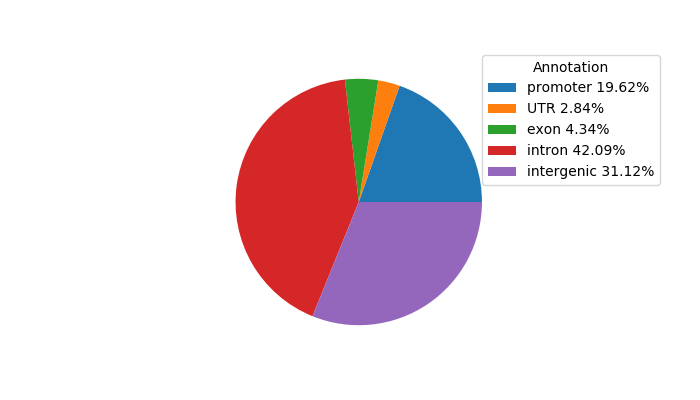
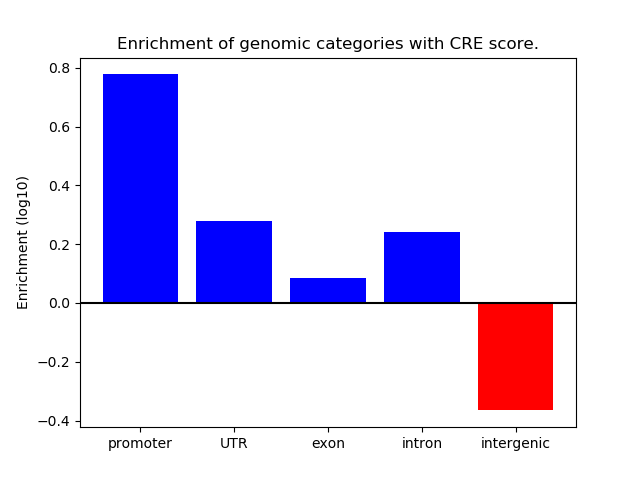
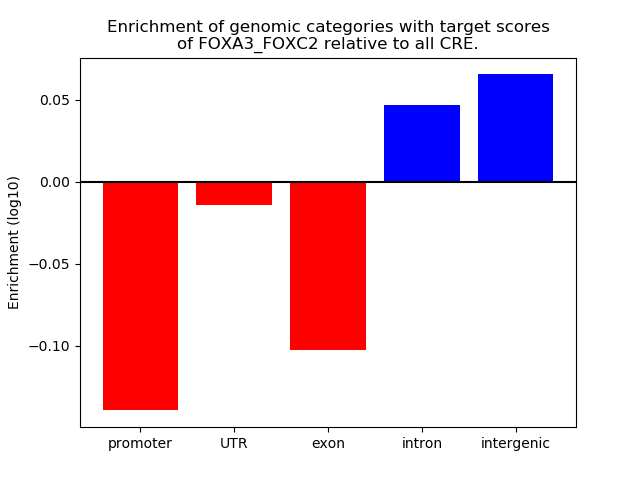
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
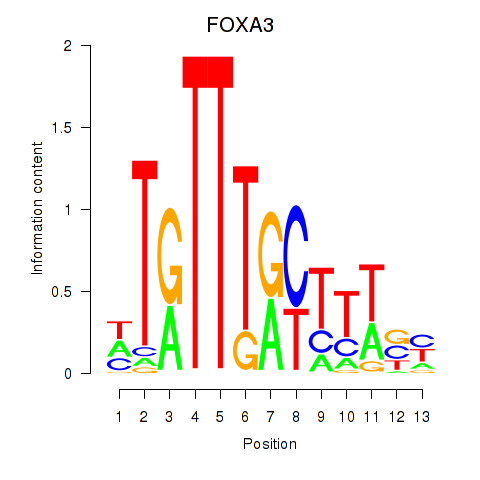
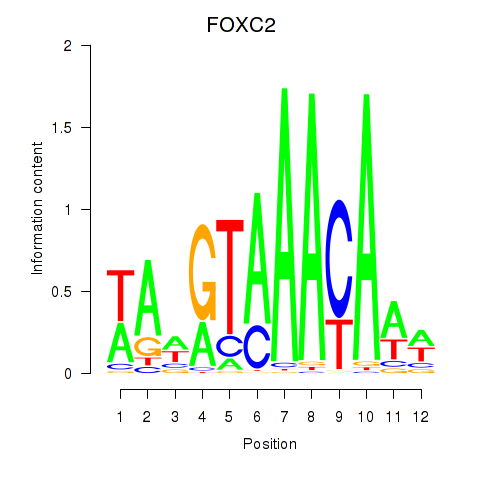
Results for FOXA3_FOXC2
Z-value: 0.63
Transcription factors associated with FOXA3_FOXC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA3
|
ENSG00000170608.2 | forkhead box A3 |
|
FOXC2
|
ENSG00000176692.4 | forkhead box C2 |
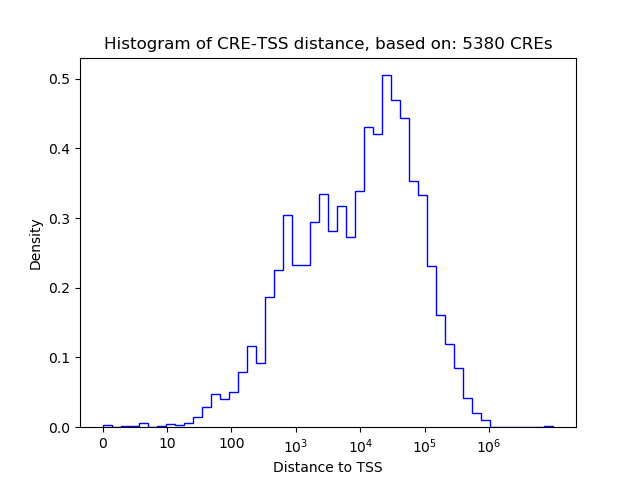
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_46367107_46367258 | FOXA3 | 65 | 0.893066 | -0.50 | 1.7e-01 | Click! |
| chr16_86597596_86598086 | FOXC2 | 3016 | 0.200096 | 0.83 | 6.1e-03 | Click! |
| chr16_86599673_86599849 | FOXC2 | 1096 | 0.389888 | 0.59 | 9.6e-02 | Click! |
| chr16_86599475_86599642 | FOXC2 | 1299 | 0.343027 | 0.53 | 1.4e-01 | Click! |
| chr16_86599945_86600151 | FOXC2 | 809 | 0.484287 | 0.50 | 1.7e-01 | Click! |
| chr16_86597297_86597487 | FOXC2 | 3465 | 0.188803 | 0.48 | 1.9e-01 | Click! |
Activity of the FOXA3_FOXC2 motif across conditions
Conditions sorted by the z-value of the FOXA3_FOXC2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
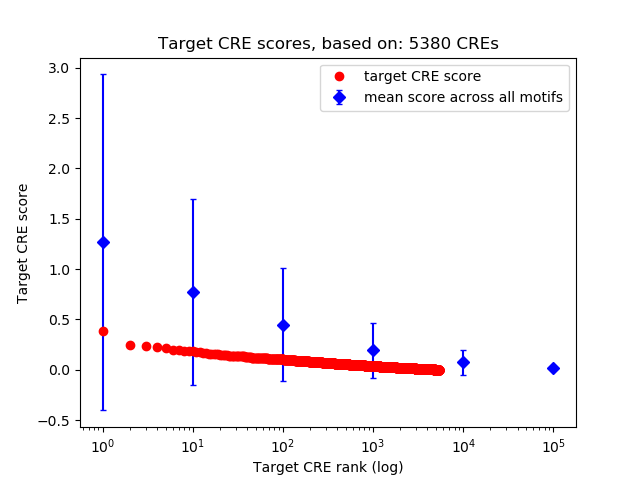
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_107738861_107739281 | 0.39 |
OXR1 |
oxidation resistance 1 |
658 |
0.71 |
| chr11_57529774_57530544 | 0.25 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
869 |
0.5 |
| chr3_167811230_167811432 | 0.23 |
GOLIM4 |
golgi integral membrane protein 4 |
1801 |
0.51 |
| chr8_15398472_15398721 | 0.23 |
TUSC3 |
tumor suppressor candidate 3 |
804 |
0.72 |
| chr6_44510355_44510599 | 0.22 |
ENSG00000266619 |
. |
107099 |
0.06 |
| chr15_96869491_96869741 | 0.20 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
449 |
0.57 |
| chr15_55574890_55575291 | 0.20 |
RAB27A |
RAB27A, member RAS oncogene family |
6911 |
0.2 |
| chr7_25109217_25109438 | 0.19 |
CYCS |
cytochrome c, somatic |
55541 |
0.14 |
| chr5_72746721_72747193 | 0.19 |
FOXD1 |
forkhead box D1 |
2605 |
0.29 |
| chr9_120466546_120467060 | 0.19 |
TLR4 |
toll-like receptor 4 |
153 |
0.96 |
| chr7_19152184_19152434 | 0.18 |
AC003986.6 |
|
212 |
0.92 |
| chr13_33803329_33803516 | 0.18 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
23279 |
0.2 |
| chr6_116833589_116833880 | 0.17 |
TRAPPC3L |
trafficking protein particle complex 3-like |
228 |
0.83 |
| chr5_52098501_52098716 | 0.16 |
CTD-2288O8.1 |
|
14748 |
0.19 |
| chr21_28214648_28215240 | 0.16 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
393 |
0.9 |
| chr14_75744052_75744985 | 0.16 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
959 |
0.51 |
| chr9_128235766_128235975 | 0.16 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
10910 |
0.23 |
| chrX_10737482_10738236 | 0.15 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
92080 |
0.1 |
| chr20_9049623_9049963 | 0.15 |
PLCB4 |
phospholipase C, beta 4 |
45 |
0.98 |
| chr2_160470923_160471074 | 0.15 |
AC009506.1 |
|
809 |
0.5 |
| chr14_33409166_33409451 | 0.15 |
NPAS3 |
neuronal PAS domain protein 3 |
785 |
0.79 |
| chr3_132036571_132036865 | 0.15 |
ACPP |
acid phosphatase, prostate |
467 |
0.84 |
| chr21_36337524_36338023 | 0.14 |
RUNX1 |
runt-related transcription factor 1 |
75686 |
0.12 |
| chr7_48284393_48284544 | 0.14 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
73411 |
0.12 |
| chr20_9048816_9049419 | 0.14 |
PLCB4 |
phospholipase C, beta 4 |
293 |
0.9 |
| chr3_100711576_100712203 | 0.14 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
408 |
0.89 |
| chr7_94025115_94025296 | 0.14 |
COL1A2 |
collagen, type I, alpha 2 |
1332 |
0.57 |
| chr10_112411260_112411724 | 0.14 |
RBM20 |
RNA binding motif protein 20 |
7337 |
0.16 |
| chr10_15126072_15126273 | 0.14 |
ACBD7 |
acyl-CoA binding domain containing 7 |
4603 |
0.18 |
| chr19_45995836_45996758 | 0.14 |
RTN2 |
reticulon 2 |
228 |
0.87 |
| chr7_19308580_19308778 | 0.14 |
FERD3L |
Fer3-like bHLH transcription factor |
123635 |
0.05 |
| chrX_10919281_10919504 | 0.14 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
67619 |
0.14 |
| chr1_158969050_158969253 | 0.13 |
IFI16 |
interferon, gamma-inducible protein 16 |
607 |
0.76 |
| chr2_190041307_190041749 | 0.13 |
COL5A2 |
collagen, type V, alpha 2 |
3077 |
0.31 |
| chr6_123039012_123039239 | 0.13 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
418 |
0.87 |
| chr6_116719287_116719943 | 0.13 |
DSE |
dermatan sulfate epimerase |
27505 |
0.14 |
| chr12_64773878_64774029 | 0.13 |
C12orf56 |
chromosome 12 open reading frame 56 |
10353 |
0.17 |
| chr12_32688080_32688676 | 0.13 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
1120 |
0.61 |
| chr6_152085356_152085807 | 0.13 |
ESR1 |
estrogen receptor 1 |
39985 |
0.18 |
| chr6_151648337_151648657 | 0.13 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
1674 |
0.27 |
| chr8_108509086_108509237 | 0.12 |
ANGPT1 |
angiopoietin 1 |
1077 |
0.7 |
| chr4_109542934_109543085 | 0.12 |
RPL34 |
ribosomal protein L34 |
1197 |
0.48 |
| chr5_59109781_59109932 | 0.12 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
45385 |
0.2 |
| chr20_1783216_1783652 | 0.12 |
SIRPA |
signal-regulatory protein alpha |
91720 |
0.07 |
| chr4_16954347_16954651 | 0.12 |
LDB2 |
LIM domain binding 2 |
54067 |
0.18 |
| chr9_12882418_12882702 | 0.12 |
ENSG00000222658 |
. |
2760 |
0.36 |
| chr5_59893443_59893927 | 0.12 |
DEPDC1B |
DEP domain containing 1B |
102276 |
0.07 |
| chr2_68465061_68465212 | 0.12 |
ENSG00000216115 |
. |
8078 |
0.14 |
| chr1_196620715_196621368 | 0.12 |
CFH |
complement factor H |
33 |
0.98 |
| chr3_133461579_133461730 | 0.12 |
TF |
transferrin |
3146 |
0.23 |
| chr1_36275039_36275190 | 0.12 |
AGO4 |
argonaute RISC catalytic component 4 |
1341 |
0.43 |
| chr16_4104228_4104558 | 0.12 |
RP11-462G12.4 |
|
22380 |
0.19 |
| chr18_65983753_65984295 | 0.12 |
TMX3 |
thioredoxin-related transmembrane protein 3 |
398270 |
0.01 |
| chr4_67430868_67431028 | 0.12 |
ENSG00000221563 |
. |
288406 |
0.01 |
| chr11_16023597_16024065 | 0.12 |
CTD-3096P4.1 |
|
20905 |
0.28 |
| chr2_150418995_150419256 | 0.12 |
AC144449.1 |
|
24585 |
0.19 |
| chr4_123795073_123795451 | 0.12 |
RP11-170N16.3 |
|
10605 |
0.18 |
| chr7_134464463_134465343 | 0.12 |
CALD1 |
caldesmon 1 |
474 |
0.89 |
| chr3_120168709_120169860 | 0.12 |
FSTL1 |
follistatin-like 1 |
554 |
0.84 |
| chr6_28175094_28175274 | 0.12 |
ZSCAN9 |
zinc finger and SCAN domain containing 9 |
17480 |
0.12 |
| chr4_87503159_87503484 | 0.12 |
MAPK10 |
mitogen-activated protein kinase 10 |
11942 |
0.21 |
| chr5_88030067_88030282 | 0.12 |
CTC-467M3.2 |
|
41706 |
0.16 |
| chr4_81392876_81393298 | 0.12 |
C4orf22 |
chromosome 4 open reading frame 22 |
136164 |
0.05 |
| chr2_157323604_157323795 | 0.11 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
6385 |
0.22 |
| chr5_90515283_90515434 | 0.11 |
ENSG00000199643 |
. |
51186 |
0.18 |
| chr2_145268671_145269078 | 0.11 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
6241 |
0.26 |
| chr6_8083977_8084191 | 0.11 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
13460 |
0.22 |
| chr12_81472525_81472676 | 0.11 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
764 |
0.72 |
| chr6_98074983_98075489 | 0.11 |
MMS22L |
MMS22-like, DNA repair protein |
344184 |
0.01 |
| chr5_67294562_67294875 | 0.11 |
ENSG00000223149 |
. |
31445 |
0.24 |
| chr10_4285552_4285907 | 0.11 |
ENSG00000207124 |
. |
271415 |
0.02 |
| chr7_80928308_80928745 | 0.11 |
AC005008.2 |
Uncharacterized protein |
111420 |
0.08 |
| chr1_158087708_158088406 | 0.11 |
KIRREL |
kin of IRRE like (Drosophila) |
31587 |
0.18 |
| chr15_77710162_77710461 | 0.11 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
2131 |
0.32 |
| chr18_32290997_32291170 | 0.11 |
DTNA |
dystrobrevin, alpha |
822 |
0.73 |
| chr17_60868253_60868692 | 0.11 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
15529 |
0.23 |
| chr18_67964238_67964389 | 0.11 |
SOCS6 |
suppressor of cytokine signaling 6 |
478 |
0.85 |
| chr5_141980216_141980760 | 0.11 |
AC005592.2 |
|
19075 |
0.18 |
| chr7_30516381_30516532 | 0.11 |
NOD1 |
nucleotide-binding oligomerization domain containing 1 |
1796 |
0.37 |
| chr1_243414977_243415342 | 0.11 |
CEP170 |
centrosomal protein 170kDa |
1463 |
0.39 |
| chr7_151328064_151328292 | 0.11 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
1261 |
0.55 |
| chr15_61044545_61044696 | 0.11 |
RP11-554D20.1 |
|
11485 |
0.22 |
| chr3_137801110_137801644 | 0.11 |
DZIP1L |
DAZ interacting zinc finger protein 1-like |
14895 |
0.2 |
| chr11_10627928_10628217 | 0.11 |
MRVI1-AS1 |
MRVI1 antisense RNA 1 |
13328 |
0.17 |
| chr5_95595176_95595327 | 0.11 |
ENSG00000206997 |
. |
49320 |
0.17 |
| chr6_141884450_141884959 | 0.11 |
ENSG00000222764 |
. |
77155 |
0.13 |
| chr12_23088184_23088587 | 0.11 |
ETNK1 |
ethanolamine kinase 1 |
310094 |
0.01 |
| chr5_82770538_82771175 | 0.11 |
VCAN |
versican |
3112 |
0.37 |
| chr2_216296824_216297216 | 0.10 |
FN1 |
fibronectin 1 |
3770 |
0.26 |
| chr10_4541408_4541559 | 0.10 |
ENSG00000207124 |
. |
15661 |
0.3 |
| chr21_40203900_40204051 | 0.10 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
22415 |
0.24 |
| chr3_162003818_162003969 | 0.10 |
OTOL1 |
otolin 1 |
789297 |
0.0 |
| chr11_61890885_61891204 | 0.10 |
INCENP |
inner centromere protein antigens 135/155kDa |
401 |
0.85 |
| chr18_37421234_37421672 | 0.10 |
ENSG00000212354 |
. |
162867 |
0.04 |
| chr12_32687758_32688027 | 0.10 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
634 |
0.8 |
| chr1_202428982_202429290 | 0.10 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
2735 |
0.3 |
| chr8_2094260_2094411 | 0.10 |
MYOM2 |
myomesin 2 |
101151 |
0.08 |
| chr9_97951408_97952029 | 0.10 |
ENSG00000238746 |
. |
16838 |
0.19 |
| chrX_15934152_15934303 | 0.10 |
ENSG00000200566 |
. |
199 |
0.96 |
| chr1_183791724_183791999 | 0.10 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
17571 |
0.24 |
| chr4_159857577_159857754 | 0.10 |
ENSG00000221314 |
. |
63777 |
0.11 |
| chr2_106260790_106261225 | 0.10 |
NCK2 |
NCK adaptor protein 2 |
100347 |
0.08 |
| chr9_112513666_112514004 | 0.10 |
RP11-406O23.2 |
|
20488 |
0.16 |
| chr6_128828944_128829440 | 0.10 |
RP1-86D1.4 |
|
2922 |
0.22 |
| chr2_180427189_180427340 | 0.10 |
ZNF385B |
zinc finger protein 385B |
51 |
0.99 |
| chr10_126843486_126843769 | 0.10 |
CTBP2 |
C-terminal binding protein 2 |
3658 |
0.35 |
| chr9_16626345_16626809 | 0.10 |
RP11-62F24.1 |
|
903 |
0.71 |
| chr3_36954201_36954545 | 0.10 |
TRANK1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
32175 |
0.17 |
| chr9_935405_935889 | 0.10 |
DMRT3 |
doublesex and mab-3 related transcription factor 3 |
41317 |
0.17 |
| chr15_99864758_99865035 | 0.10 |
AC022819.2 |
Uncharacterized protein |
5068 |
0.23 |
| chr8_48297219_48297488 | 0.10 |
SPIDR |
scaffolding protein involved in DNA repair |
55610 |
0.16 |
| chr7_90224844_90225183 | 0.10 |
AC002456.2 |
|
49 |
0.94 |
| chr6_143995099_143995343 | 0.10 |
PHACTR2 |
phosphatase and actin regulator 2 |
3881 |
0.32 |
| chr2_188416841_188416992 | 0.10 |
AC007319.1 |
|
1960 |
0.34 |
| chr5_124082726_124083211 | 0.10 |
ZNF608 |
zinc finger protein 608 |
648 |
0.62 |
| chr10_116639288_116639471 | 0.10 |
FAM160B1 |
family with sequence similarity 160, member B1 |
18829 |
0.24 |
| chr18_52763849_52764148 | 0.10 |
ENSG00000252437 |
. |
49878 |
0.16 |
| chr3_63908711_63908862 | 0.10 |
ATXN7 |
ataxin 7 |
10511 |
0.18 |
| chr6_45523679_45523878 | 0.10 |
ENSG00000252738 |
. |
90063 |
0.09 |
| chr18_55890673_55891004 | 0.10 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
2035 |
0.39 |
| chr9_22883230_22883381 | 0.10 |
ENSG00000265572 |
. |
69797 |
0.14 |
| chr17_68164439_68165051 | 0.10 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
69 |
0.94 |
| chr15_53072694_53072932 | 0.10 |
ONECUT1 |
one cut homeobox 1 |
567 |
0.84 |
| chr9_22007341_22007594 | 0.09 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
1485 |
0.34 |
| chr12_91575790_91576058 | 0.09 |
DCN |
decorin |
505 |
0.87 |
| chr7_72991877_72992028 | 0.09 |
TBL2 |
transducin (beta)-like 2 |
714 |
0.6 |
| chr14_74551356_74551507 | 0.09 |
LIN52 |
lin-52 homolog (C. elegans) |
68 |
0.72 |
| chr2_127983002_127983353 | 0.09 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
5523 |
0.25 |
| chr7_3632163_3632314 | 0.09 |
AC011284.3 |
|
51178 |
0.18 |
| chr3_171685206_171685443 | 0.09 |
FNDC3B |
fibronectin type III domain containing 3B |
72094 |
0.1 |
| chr18_72880123_72880274 | 0.09 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
37270 |
0.19 |
| chr1_87254203_87254481 | 0.09 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
83764 |
0.08 |
| chr10_4284322_4284712 | 0.09 |
ENSG00000207124 |
. |
272627 |
0.02 |
| chr17_38604976_38605127 | 0.09 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
5338 |
0.15 |
| chr5_38550048_38550208 | 0.09 |
LIFR |
leukemia inhibitory factor receptor alpha |
6610 |
0.18 |
| chr6_140525151_140525313 | 0.09 |
ENSG00000263514 |
. |
1157 |
0.62 |
| chr6_71972702_71972953 | 0.09 |
RP11-154D6.1 |
|
21808 |
0.19 |
| chr6_112823116_112823311 | 0.09 |
ENSG00000239095 |
. |
25285 |
0.2 |
| chr21_43316610_43316761 | 0.09 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
5076 |
0.15 |
| chr4_129557030_129557181 | 0.09 |
ENSG00000238802 |
. |
120076 |
0.06 |
| chr3_120361749_120361900 | 0.09 |
HGD |
homogentisate 1,2-dioxygenase |
3358 |
0.3 |
| chr7_143581670_143581927 | 0.09 |
FAM115A |
family with sequence similarity 115, member A |
665 |
0.7 |
| chr2_179032632_179032866 | 0.09 |
ENSG00000223096 |
. |
25474 |
0.16 |
| chr10_28015309_28015460 | 0.09 |
MKX |
mohawk homeobox |
17090 |
0.2 |
| chr3_15370902_15371053 | 0.09 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
3090 |
0.19 |
| chr3_138594988_138595139 | 0.09 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
41283 |
0.15 |
| chr6_82496917_82497068 | 0.09 |
ENSG00000206886 |
. |
23251 |
0.2 |
| chr8_96820368_96820519 | 0.09 |
ENSG00000223297 |
. |
129300 |
0.06 |
| chr1_98369641_98369792 | 0.09 |
DPYD |
dihydropyrimidine dehydrogenase |
16837 |
0.28 |
| chr6_45296305_45296528 | 0.09 |
RUNX2 |
runt-related transcription factor 2 |
48 |
0.98 |
| chr7_74293974_74294125 | 0.09 |
STAG3L2 |
stromal antigen 3-like 2 (pseudogene) |
10469 |
0.18 |
| chr10_123469524_123469675 | 0.09 |
RP11-78A18.2 |
|
26219 |
0.24 |
| chr3_64199959_64200146 | 0.09 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
11079 |
0.21 |
| chr9_128484739_128484890 | 0.09 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
15301 |
0.23 |
| chr14_79747812_79748200 | 0.09 |
NRXN3 |
neurexin 3 |
1757 |
0.44 |
| chr4_134071254_134071890 | 0.09 |
PCDH10 |
protocadherin 10 |
1102 |
0.7 |
| chr4_57276664_57276969 | 0.09 |
AC068620.1 |
Uncharacterized protein |
134 |
0.95 |
| chr10_17278277_17278428 | 0.09 |
RP11-124N14.3 |
|
1520 |
0.34 |
| chr1_186472613_186472786 | 0.09 |
PDC |
phosducin |
42445 |
0.15 |
| chr1_81703498_81704139 | 0.09 |
ENSG00000223026 |
. |
13656 |
0.28 |
| chr4_186577993_186578477 | 0.09 |
SORBS2 |
sorbin and SH3 domain containing 2 |
112 |
0.97 |
| chr11_13204755_13204972 | 0.09 |
ARNTL |
aryl hydrocarbon receptor nuclear translocator-like |
93336 |
0.09 |
| chr6_150177526_150177677 | 0.09 |
RP11-350J20.12 |
|
4003 |
0.14 |
| chr15_39881812_39881998 | 0.09 |
CTD-2033D15.1 |
|
4527 |
0.21 |
| chr2_201473941_201474433 | 0.09 |
AOX1 |
aldehyde oxidase 1 |
21290 |
0.2 |
| chr12_75876585_75876939 | 0.09 |
GLIPR1 |
GLI pathogenesis-related 1 |
1778 |
0.34 |
| chr9_89808674_89808967 | 0.09 |
C9orf170 |
chromosome 9 open reading frame 170 |
45261 |
0.18 |
| chr20_1103594_1103745 | 0.09 |
PSMF1 |
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
4389 |
0.25 |
| chr6_72205767_72206133 | 0.09 |
ENSG00000212099 |
. |
42729 |
0.17 |
| chr18_60774564_60774715 | 0.09 |
RP11-299P2.1 |
|
43914 |
0.17 |
| chr4_48701412_48701950 | 0.09 |
FRYL |
FRY-like |
18493 |
0.24 |
| chr4_169553568_169553728 | 0.09 |
PALLD |
palladin, cytoskeletal associated protein |
880 |
0.63 |
| chr5_115790032_115790655 | 0.09 |
CTB-118N6.3 |
|
4917 |
0.26 |
| chr1_151653168_151653448 | 0.09 |
RP11-98D18.1 |
|
20194 |
0.08 |
| chr10_54357230_54357516 | 0.09 |
RP11-556E13.1 |
|
36508 |
0.23 |
| chr2_63416100_63416251 | 0.09 |
OTX1 |
orthodenticle homeobox 1 |
138238 |
0.05 |
| chr11_86085513_86085741 | 0.09 |
CCDC81 |
coiled-coil domain containing 81 |
151 |
0.96 |
| chr1_116665777_116665975 | 0.09 |
MAB21L3 |
mab-21-like 3 (C. elegans) |
11500 |
0.27 |
| chr5_121417237_121417571 | 0.09 |
LOX |
lysyl oxidase |
3424 |
0.28 |
| chr9_14098246_14098397 | 0.08 |
NFIB |
nuclear factor I/B |
82470 |
0.11 |
| chr5_74634021_74634424 | 0.08 |
HMGCR |
3-hydroxy-3-methylglutaryl-CoA reductase |
264 |
0.89 |
| chr3_25471806_25472309 | 0.08 |
RARB |
retinoic acid receptor, beta |
2255 |
0.41 |
| chr4_115282272_115282768 | 0.08 |
UGT8 |
UDP glycosyltransferase 8 |
237091 |
0.02 |
| chr15_48959812_48960090 | 0.08 |
FBN1 |
fibrillin 1 |
21905 |
0.23 |
| chr11_60926256_60926465 | 0.08 |
VPS37C |
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
2248 |
0.27 |
| chr1_119529921_119530106 | 0.08 |
TBX15 |
T-box 15 |
415 |
0.9 |
| chr7_82057700_82058025 | 0.08 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
15169 |
0.3 |
| chr3_105584255_105584406 | 0.08 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
3557 |
0.4 |
| chr2_28189640_28189791 | 0.08 |
ENSG00000265321 |
. |
29519 |
0.18 |
| chr4_96455327_96455478 | 0.08 |
UNC5C |
unc-5 homolog C (C. elegans) |
14721 |
0.24 |
| chr6_19465542_19465832 | 0.08 |
ENSG00000201523 |
. |
27173 |
0.27 |
| chr2_224445554_224445715 | 0.08 |
SCG2 |
secretogranin II |
21368 |
0.28 |
| chr15_96883311_96883827 | 0.08 |
ENSG00000222651 |
. |
7079 |
0.16 |
| chr12_115174256_115174747 | 0.08 |
ENSG00000252459 |
. |
1008 |
0.64 |
| chr4_138585410_138585827 | 0.08 |
PCDH18 |
protocadherin 18 |
131970 |
0.06 |
| chr12_94132814_94133007 | 0.08 |
RP11-887P2.5 |
|
1311 |
0.51 |
| chr3_43337483_43337634 | 0.08 |
SNRK |
SNF related kinase |
9480 |
0.18 |
| chr2_128491676_128491827 | 0.08 |
SFT2D3 |
SFT2 domain containing 3 |
33154 |
0.13 |
| chr8_9409853_9410128 | 0.08 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
3434 |
0.27 |
| chr4_186696107_186696537 | 0.08 |
SORBS2 |
sorbin and SH3 domain containing 2 |
108 |
0.98 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.0 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0060841 | venous blood vessel morphogenesis(GO:0048845) venous blood vessel development(GO:0060841) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.0 | GO:0042321 | negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.0 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.0 | GO:0072071 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0015802 | basic amino acid transport(GO:0015802) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |