Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
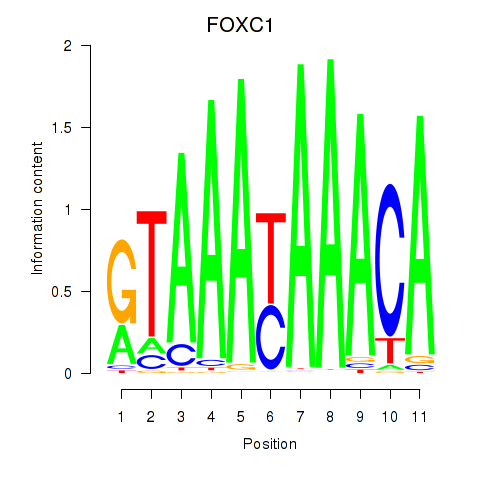
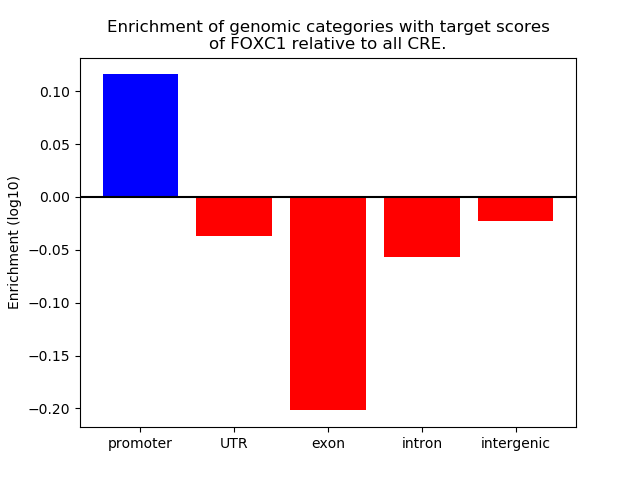
Results for FOXC1
Z-value: 0.81
Transcription factors associated with FOXC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXC1
|
ENSG00000054598.5 | forkhead box C1 |
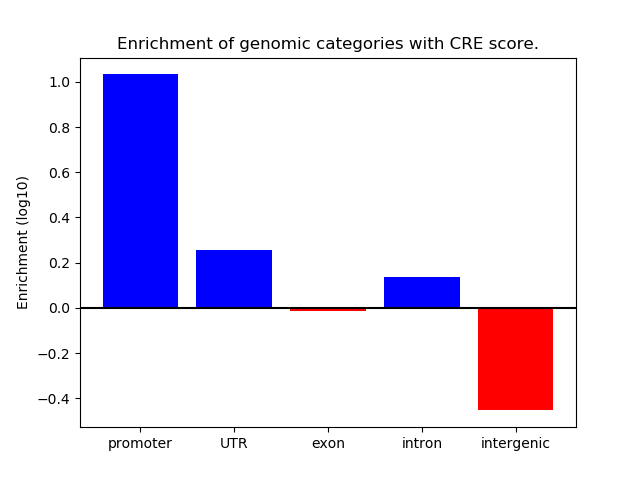
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_1885704_1885932 | FOXC1 | 275137 | 0.014966 | 0.70 | 3.7e-02 | Click! |
| chr6_1703862_1704013 | FOXC1 | 93256 | 0.093857 | -0.58 | 1.0e-01 | Click! |
| chr6_1617814_1617965 | FOXC1 | 7208 | 0.314162 | 0.57 | 1.1e-01 | Click! |
| chr6_1615364_1615572 | FOXC1 | 4787 | 0.336813 | 0.56 | 1.1e-01 | Click! |
| chr6_1610269_1610509 | FOXC1 | 292 | 0.947549 | 0.56 | 1.2e-01 | Click! |
Activity of the FOXC1 motif across conditions
Conditions sorted by the z-value of the FOXC1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
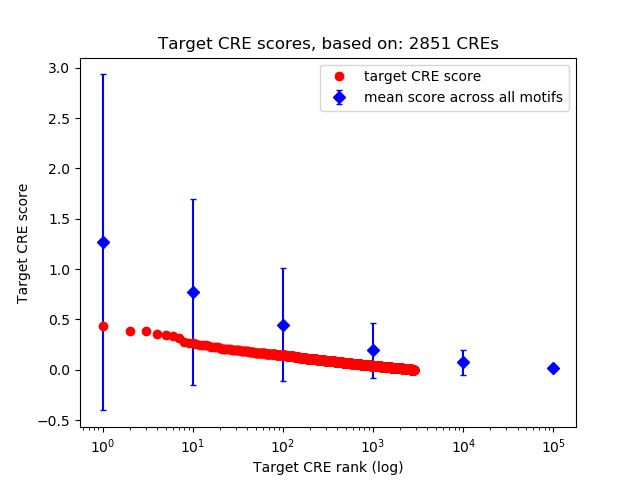
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_16826274_16826456 | 0.43 |
TXLNG |
taxilin gamma |
21810 |
0.16 |
| chr7_6521360_6521521 | 0.39 |
FLJ20306 |
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
1482 |
0.28 |
| chr11_82828119_82828301 | 0.39 |
PCF11 |
PCF11 cleavage and polyadenylation factor subunit |
39820 |
0.12 |
| chr11_3400503_3400805 | 0.35 |
ZNF195 |
zinc finger protein 195 |
206 |
0.92 |
| chr8_27631557_27631733 | 0.34 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
258 |
0.66 |
| chr8_77911562_77912296 | 0.33 |
PEX2 |
peroxisomal biogenesis factor 2 |
533 |
0.85 |
| chr9_71590240_71590555 | 0.32 |
PRKACG |
protein kinase, cAMP-dependent, catalytic, gamma |
38642 |
0.15 |
| chr7_84154352_84154503 | 0.28 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
32387 |
0.25 |
| chr11_92930195_92930700 | 0.27 |
SLC36A4 |
solute carrier family 36 (proton/amino acid symporter), member 4 |
174 |
0.97 |
| chr1_98369641_98369792 | 0.26 |
DPYD |
dihydropyrimidine dehydrogenase |
16837 |
0.28 |
| chr6_1612835_1613496 | 0.26 |
FOXC1 |
forkhead box C1 |
2484 |
0.42 |
| chr11_119076374_119076525 | 0.25 |
CBL |
Cbl proto-oncogene, E3 ubiquitin protein ligase |
303 |
0.81 |
| chr12_75876585_75876939 | 0.25 |
GLIPR1 |
GLI pathogenesis-related 1 |
1778 |
0.34 |
| chr4_129557030_129557181 | 0.25 |
ENSG00000238802 |
. |
120076 |
0.06 |
| chr11_46615706_46615915 | 0.24 |
AMBRA1 |
autophagy/beclin-1 regulator 1 |
135 |
0.96 |
| chr10_50649676_50649900 | 0.23 |
RP11-123B3.2 |
|
22465 |
0.17 |
| chr12_72667945_72668096 | 0.23 |
ENSG00000236333 |
. |
667 |
0.5 |
| chr14_31879360_31879511 | 0.23 |
HEATR5A |
HEAT repeat containing 5A |
7254 |
0.17 |
| chr8_13730073_13730224 | 0.23 |
RP11-145O15.3 |
|
268029 |
0.02 |
| chr15_76629589_76629763 | 0.22 |
ISL2 |
ISL LIM homeobox 2 |
611 |
0.74 |
| chrX_15934152_15934303 | 0.21 |
ENSG00000200566 |
. |
199 |
0.96 |
| chr15_91955379_91955670 | 0.21 |
SV2B |
synaptic vesicle glycoprotein 2B |
186424 |
0.03 |
| chr6_116833589_116833880 | 0.21 |
TRAPPC3L |
trafficking protein particle complex 3-like |
228 |
0.83 |
| chr19_2095175_2095486 | 0.21 |
MOB3A |
MOB kinase activator 3A |
939 |
0.33 |
| chr2_113012718_113012869 | 0.21 |
ZC3H8 |
zinc finger CCCH-type containing 8 |
105 |
0.97 |
| chr10_91061669_91062419 | 0.20 |
IFIT2 |
interferon-induced protein with tetratricopeptide repeats 2 |
332 |
0.85 |
| chrY_9925657_9925808 | 0.20 |
ENSG00000251766 |
. |
2405 |
0.28 |
| chr16_2570012_2570182 | 0.20 |
AMDHD2 |
amidohydrolase domain containing 2 |
261 |
0.71 |
| chr11_46141542_46141818 | 0.20 |
PHF21A |
PHD finger protein 21A |
267 |
0.92 |
| chrX_18456912_18457063 | 0.20 |
CDKL5 |
cyclin-dependent kinase-like 5 |
3321 |
0.31 |
| chr11_104576737_104577140 | 0.20 |
CASP12 |
caspase 12 (gene/pseudogene) |
192203 |
0.03 |
| chr1_28832516_28832718 | 0.19 |
RCC1 |
regulator of chromosome condensation 1 |
48 |
0.94 |
| chr10_8203120_8203271 | 0.19 |
GATA3 |
GATA binding protein 3 |
106426 |
0.08 |
| chr2_72376882_72377179 | 0.19 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
1863 |
0.53 |
| chr5_158407500_158407684 | 0.19 |
CTD-2363C16.1 |
|
2422 |
0.33 |
| chr6_2876062_2876267 | 0.19 |
ENSG00000266750 |
. |
21823 |
0.15 |
| chr4_10691726_10691877 | 0.19 |
CLNK |
cytokine-dependent hematopoietic cell linker |
5312 |
0.35 |
| chr1_39324368_39325146 | 0.19 |
RRAGC |
Ras-related GTP binding C |
738 |
0.44 |
| chr5_56124620_56124771 | 0.19 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
13294 |
0.17 |
| chr11_15919049_15919200 | 0.18 |
CTD-3096P4.1 |
|
125612 |
0.06 |
| chr7_147748643_147748794 | 0.18 |
CNTNAP2 |
contactin associated protein-like 2 |
82058 |
0.1 |
| chr19_4114935_4115086 | 0.18 |
MAP2K2 |
mitogen-activated protein kinase kinase 2 |
9104 |
0.12 |
| chr4_169075013_169075164 | 0.18 |
RP11-310I9.1 |
|
40606 |
0.17 |
| chr18_21032782_21033062 | 0.18 |
RIOK3 |
RIO kinase 3 |
135 |
0.95 |
| chr2_61372387_61372598 | 0.18 |
C2orf74 |
chromosome 2 open reading frame 74 |
249 |
0.92 |
| chr5_17255187_17255338 | 0.18 |
ENSG00000252908 |
. |
14542 |
0.19 |
| chr6_132833581_132834005 | 0.18 |
STX7 |
syntaxin 7 |
408 |
0.85 |
| chr14_52116783_52117087 | 0.17 |
FRMD6 |
FERM domain containing 6 |
1641 |
0.39 |
| chr6_156086673_156086824 | 0.17 |
ENSG00000238789 |
. |
110588 |
0.07 |
| chr11_57529774_57530544 | 0.17 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
869 |
0.5 |
| chr4_7937771_7938061 | 0.17 |
AC097381.1 |
|
2812 |
0.26 |
| chr8_126444680_126445315 | 0.17 |
TRIB1 |
tribbles pseudokinase 1 |
546 |
0.81 |
| chr12_91574306_91575101 | 0.17 |
DCN |
decorin |
50 |
0.99 |
| chr19_19932661_19932812 | 0.17 |
ZNF506 |
zinc finger protein 506 |
176 |
0.93 |
| chr5_82672808_82672959 | 0.17 |
VCAN |
versican |
94401 |
0.09 |
| chr12_77626162_77626313 | 0.17 |
ENSG00000238769 |
. |
69432 |
0.14 |
| chr14_61190612_61190839 | 0.17 |
SIX4 |
SIX homeobox 4 |
127 |
0.97 |
| chr19_42985275_42985426 | 0.17 |
ENSG00000253048 |
. |
8323 |
0.14 |
| chr14_75744052_75744985 | 0.16 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
959 |
0.51 |
| chr7_116775065_116775216 | 0.16 |
ST7-AS2 |
ST7 antisense RNA 2 |
10472 |
0.23 |
| chr14_37463384_37463535 | 0.16 |
ENSG00000266327 |
. |
41863 |
0.18 |
| chr7_24608273_24608424 | 0.16 |
MPP6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
4539 |
0.29 |
| chr20_17005032_17005183 | 0.16 |
ENSG00000212165 |
. |
41429 |
0.2 |
| chr10_63855843_63855994 | 0.16 |
ENSG00000221094 |
. |
20056 |
0.23 |
| chr3_79778551_79778702 | 0.16 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
38339 |
0.23 |
| chr13_24841248_24841663 | 0.16 |
SPATA13 |
spermatogenesis associated 13 |
3371 |
0.21 |
| chr5_54157434_54157674 | 0.16 |
ENSG00000221073 |
. |
8778 |
0.26 |
| chr1_64977055_64977206 | 0.16 |
CACHD1 |
cache domain containing 1 |
40655 |
0.18 |
| chr5_369067_369218 | 0.16 |
AHRR |
aryl-hydrocarbon receptor repressor |
25499 |
0.15 |
| chr18_47793998_47794618 | 0.16 |
CCDC11 |
coiled-coil domain containing 11 |
1416 |
0.36 |
| chr2_216293163_216293314 | 0.16 |
FN1 |
fibronectin 1 |
7552 |
0.23 |
| chr8_59466247_59466777 | 0.16 |
SDCBP |
syndecan binding protein (syntenin) |
698 |
0.76 |
| chr7_134287566_134287824 | 0.16 |
AKR1B15 |
aldo-keto reductase family 1, member B15 |
37462 |
0.17 |
| chr1_28879171_28879359 | 0.16 |
TRNAU1AP |
tRNA selenocysteine 1 associated protein 1 |
332 |
0.8 |
| chr15_64923415_64923566 | 0.16 |
ENSG00000207223 |
. |
21597 |
0.14 |
| chr9_128484739_128484890 | 0.16 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
15301 |
0.23 |
| chr8_42005635_42005786 | 0.16 |
RP11-589C21.5 |
|
4571 |
0.19 |
| chr22_17697616_17697767 | 0.16 |
CECR1 |
cat eye syndrome chromosome region, candidate 1 |
2575 |
0.29 |
| chr13_27825183_27825382 | 0.15 |
RPL21 |
ribosomal protein L21 |
164 |
0.92 |
| chr19_16347709_16347860 | 0.15 |
AP1M1 |
adaptor-related protein complex 1, mu 1 subunit |
9368 |
0.15 |
| chr6_159458069_159458220 | 0.15 |
TAGAP |
T-cell activation RhoGTPase activating protein |
7906 |
0.19 |
| chr3_187984346_187984497 | 0.15 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
26769 |
0.24 |
| chr10_17826229_17826380 | 0.15 |
MRC1L1 |
cDNA FLJ56855, highly similar to Macrophage mannose receptor 1 |
25058 |
0.16 |
| chr20_5550410_5550561 | 0.15 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
5669 |
0.27 |
| chr10_28822829_28823308 | 0.15 |
WAC |
WW domain containing adaptor with coiled-coil |
16 |
0.97 |
| chr7_93926282_93926700 | 0.15 |
COL1A2 |
collagen, type I, alpha 2 |
97382 |
0.08 |
| chr5_55339622_55339773 | 0.15 |
ENSG00000238326 |
. |
43248 |
0.11 |
| chr5_37370451_37371178 | 0.15 |
NUP155 |
nucleoporin 155kDa |
73 |
0.98 |
| chr7_70773511_70773662 | 0.15 |
ENSG00000265878 |
. |
844 |
0.58 |
| chr5_15575280_15575431 | 0.15 |
CTD-2350J17.1 |
|
32003 |
0.21 |
| chr1_205144953_205145111 | 0.15 |
DSTYK |
dual serine/threonine and tyrosine protein kinase |
35662 |
0.13 |
| chr14_90863482_90863819 | 0.15 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
250 |
0.92 |
| chr3_31814428_31814579 | 0.15 |
OSBPL10-AS1 |
OSBPL10 antisense RNA 1 |
56020 |
0.14 |
| chr2_152146458_152146912 | 0.15 |
NMI |
N-myc (and STAT) interactor |
114 |
0.97 |
| chrX_80458564_80458878 | 0.15 |
SH3BGRL |
SH3 domain binding glutamic acid-rich protein like |
1279 |
0.36 |
| chr1_91748546_91748727 | 0.15 |
ENSG00000206817 |
. |
21454 |
0.25 |
| chr12_56435716_56435907 | 0.15 |
RPS26 |
ribosomal protein S26 |
134 |
0.6 |
| chr14_23584164_23584381 | 0.15 |
CEBPE |
CCAAT/enhancer binding protein (C/EBP), epsilon |
4553 |
0.11 |
| chr9_97021933_97022185 | 0.14 |
ZNF169 |
zinc finger protein 169 |
449 |
0.8 |
| chr19_10927638_10927909 | 0.14 |
ENSG00000207752 |
. |
399 |
0.76 |
| chr5_66255088_66255484 | 0.14 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
470 |
0.88 |
| chr1_109935632_109935931 | 0.14 |
SORT1 |
sortilin 1 |
198 |
0.93 |
| chr3_119921742_119921893 | 0.14 |
ENSG00000206721 |
. |
7350 |
0.22 |
| chr8_8395574_8395725 | 0.14 |
ENSG00000263616 |
. |
44516 |
0.17 |
| chr9_20144272_20144623 | 0.14 |
ENSG00000221744 |
. |
150555 |
0.04 |
| chr15_69110393_69110544 | 0.14 |
ANP32A |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
2757 |
0.31 |
| chr10_104926250_104926401 | 0.14 |
NT5C2 |
5'-nucleotidase, cytosolic II |
8414 |
0.23 |
| chr7_148843913_148844354 | 0.14 |
ZNF398 |
zinc finger protein 398 |
427 |
0.79 |
| chr8_37556330_37556485 | 0.14 |
ZNF703 |
zinc finger protein 703 |
3138 |
0.17 |
| chr3_122513057_122513432 | 0.14 |
DIRC2 |
disrupted in renal carcinoma 2 |
398 |
0.6 |
| chr6_6592584_6592735 | 0.14 |
LY86 |
lymphocyte antigen 86 |
3757 |
0.28 |
| chr6_106583373_106583524 | 0.14 |
RP1-134E15.3 |
|
35433 |
0.17 |
| chr9_120466546_120467060 | 0.14 |
TLR4 |
toll-like receptor 4 |
153 |
0.96 |
| chr6_113012091_113012242 | 0.14 |
ENSG00000252215 |
. |
158782 |
0.04 |
| chr5_179482246_179482397 | 0.14 |
RNF130 |
ring finger protein 130 |
16400 |
0.19 |
| chr1_236767249_236767530 | 0.14 |
HEATR1 |
HEAT repeat containing 1 |
399 |
0.87 |
| chr7_19308580_19308778 | 0.14 |
FERD3L |
Fer3-like bHLH transcription factor |
123635 |
0.05 |
| chr6_151597751_151597996 | 0.14 |
ENSG00000252431 |
. |
3874 |
0.18 |
| chr21_34499350_34499501 | 0.14 |
AP000290.7 |
|
29513 |
0.14 |
| chr12_124043713_124043864 | 0.14 |
ENSG00000266655 |
. |
22832 |
0.11 |
| chr5_133583428_133583579 | 0.14 |
PPP2CA |
protein phosphatase 2, catalytic subunit, alpha isozyme |
21670 |
0.14 |
| chr1_66797787_66798583 | 0.14 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
313 |
0.94 |
| chr13_30422824_30423057 | 0.14 |
UBL3 |
ubiquitin-like 3 |
1881 |
0.48 |
| chr17_42767403_42767760 | 0.14 |
CCDC43 |
coiled-coil domain containing 43 |
434 |
0.75 |
| chr5_98369155_98369504 | 0.14 |
ENSG00000200351 |
. |
96878 |
0.08 |
| chr8_92051814_92052056 | 0.14 |
TMEM55A |
transmembrane protein 55A |
1128 |
0.39 |
| chr6_79788058_79788292 | 0.13 |
PHIP |
pleckstrin homology domain interacting protein |
222 |
0.96 |
| chr2_144565732_144565883 | 0.13 |
CTD-2252P21.1 |
|
32165 |
0.19 |
| chr11_94226416_94226932 | 0.13 |
ANKRD49 |
ankyrin repeat domain 49 |
121 |
0.73 |
| chr22_17651901_17652140 | 0.13 |
CECR5 |
cat eye syndrome chromosome region, candidate 5 |
5843 |
0.17 |
| chr1_95700160_95700399 | 0.13 |
RWDD3 |
RWD domain containing 3 |
512 |
0.57 |
| chr6_26365718_26366052 | 0.13 |
ENSG00000215979 |
. |
105 |
0.79 |
| chr6_134694363_134694677 | 0.13 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
55270 |
0.15 |
| chr19_13278225_13278635 | 0.13 |
CTC-250I14.6 |
|
13423 |
0.11 |
| chr10_105595201_105595352 | 0.13 |
SH3PXD2A |
SH3 and PX domains 2A |
19888 |
0.19 |
| chr14_59106280_59106431 | 0.13 |
DACT1 |
dishevelled-binding antagonist of beta-catenin 1 |
800 |
0.74 |
| chr10_75646822_75647179 | 0.13 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
12657 |
0.12 |
| chr6_26125804_26126001 | 0.13 |
HIST1H2AC |
histone cluster 1, H2ac |
1471 |
0.17 |
| chr9_23190046_23190197 | 0.13 |
ENSG00000265572 |
. |
237019 |
0.02 |
| chr18_10743903_10744054 | 0.13 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
4520 |
0.28 |
| chr5_173318326_173318892 | 0.13 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
929 |
0.69 |
| chr1_66800579_66801197 | 0.12 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
3016 |
0.39 |
| chr10_14049890_14050132 | 0.12 |
FRMD4A |
FERM domain containing 4A |
521 |
0.82 |
| chr1_2164202_2164388 | 0.12 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
4161 |
0.15 |
| chr16_48418037_48418576 | 0.12 |
SIAH1 |
siah E3 ubiquitin protein ligase 1 |
1055 |
0.5 |
| chr15_67803077_67803228 | 0.12 |
C15orf61 |
chromosome 15 open reading frame 61 |
10254 |
0.21 |
| chr11_67142318_67142469 | 0.12 |
CLCF1 |
cardiotrophin-like cytokine factor 1 |
745 |
0.41 |
| chr4_22239606_22240002 | 0.12 |
ENSG00000238383 |
. |
111269 |
0.07 |
| chr6_56555605_56555856 | 0.12 |
DST |
dystonin |
47936 |
0.18 |
| chr1_221219794_221220024 | 0.12 |
HLX |
H2.0-like homeobox |
165325 |
0.04 |
| chr17_34891354_34891543 | 0.12 |
PIGW |
phosphatidylinositol glycan anchor biosynthesis, class W |
45 |
0.91 |
| chr1_159852065_159852216 | 0.12 |
RP11-190A12.8 |
|
15396 |
0.08 |
| chr1_150979434_150979585 | 0.12 |
FAM63A |
family with sequence similarity 63, member A |
176 |
0.86 |
| chr17_39471970_39472121 | 0.12 |
KRTAP17-1 |
keratin associated protein 17-1 |
98 |
0.92 |
| chrX_39501672_39501823 | 0.12 |
ENSG00000263730 |
. |
18723 |
0.29 |
| chr11_122568313_122568557 | 0.12 |
ENSG00000239079 |
. |
28584 |
0.19 |
| chr6_2222017_2222664 | 0.12 |
GMDS |
GDP-mannose 4,6-dehydratase |
23586 |
0.28 |
| chr5_123463412_123463801 | 0.12 |
CSNK1G3 |
casein kinase 1, gamma 3 |
539487 |
0.0 |
| chr7_92746852_92747341 | 0.12 |
SAMD9 |
sterile alpha motif domain containing 9 |
179 |
0.96 |
| chr9_127704164_127704315 | 0.12 |
GOLGA1 |
golgin A1 |
861 |
0.6 |
| chr3_15370902_15371053 | 0.12 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
3090 |
0.19 |
| chr20_42087283_42087478 | 0.12 |
SRSF6 |
serine/arginine-rich splicing factor 6 |
812 |
0.56 |
| chr2_25457035_25457186 | 0.12 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
18070 |
0.19 |
| chr22_18120777_18121028 | 0.12 |
BCL2L13 |
BCL2-like 13 (apoptosis facilitator) |
454 |
0.78 |
| chr17_56709506_56709776 | 0.12 |
ENSG00000212195 |
. |
444 |
0.75 |
| chr8_22525362_22525513 | 0.12 |
BIN3 |
bridging integrator 3 |
1189 |
0.35 |
| chr7_72772148_72772660 | 0.12 |
ENSG00000206709 |
. |
19201 |
0.13 |
| chr5_140044777_140044956 | 0.12 |
WDR55 |
WD repeat domain 55 |
605 |
0.47 |
| chr16_67222162_67222313 | 0.12 |
EXOC3L1 |
exocyst complex component 3-like 1 |
1802 |
0.13 |
| chr6_6943480_6943659 | 0.12 |
ENSG00000240936 |
. |
4439 |
0.33 |
| chr18_56168357_56168508 | 0.12 |
RP11-1151B14.4 |
|
34215 |
0.13 |
| chr1_205257217_205257368 | 0.12 |
TMCC2 |
transmembrane and coiled-coil domain family 2 |
31963 |
0.12 |
| chr20_56963544_56964018 | 0.11 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
397 |
0.89 |
| chr9_99776295_99776451 | 0.11 |
HIATL2 |
hippocampus abundant transcript-like 2 |
511 |
0.83 |
| chr12_75875359_75875792 | 0.11 |
GLIPR1 |
GLI pathogenesis-related 1 |
591 |
0.74 |
| chr12_53448318_53448482 | 0.11 |
RP11-983P16.4 |
|
178 |
0.91 |
| chrY_23354523_23354674 | 0.11 |
CYorf17 |
chromosome Y open reading frame 17 |
193648 |
0.03 |
| chr10_120789931_120790695 | 0.11 |
NANOS1 |
nanos homolog 1 (Drosophila) |
375 |
0.81 |
| chr3_11278875_11279026 | 0.11 |
HRH1 |
histamine receptor H1 |
11233 |
0.25 |
| chr19_34286123_34286306 | 0.11 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
624 |
0.82 |
| chr15_72665455_72665606 | 0.11 |
HEXA |
hexosaminidase A (alpha polypeptide) |
2655 |
0.21 |
| chr18_72880123_72880274 | 0.11 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
37270 |
0.19 |
| chr4_24975617_24975768 | 0.11 |
CCDC149 |
coiled-coil domain containing 149 |
6078 |
0.28 |
| chr12_105502043_105502544 | 0.11 |
KIAA1033 |
KIAA1033 |
801 |
0.5 |
| chr4_162498417_162498568 | 0.11 |
RP11-234O6.2 |
|
198387 |
0.03 |
| chr2_192900733_192900884 | 0.11 |
AC098617.1 |
|
102 |
0.98 |
| chr14_89796844_89796995 | 0.11 |
RP11-356K23.1 |
|
19710 |
0.18 |
| chrX_3617821_3618286 | 0.11 |
PRKX |
protein kinase, X-linked |
13596 |
0.21 |
| chr4_15704055_15704284 | 0.11 |
BST1 |
bone marrow stromal cell antigen 1 |
404 |
0.85 |
| chr1_72749102_72749377 | 0.11 |
NEGR1 |
neuronal growth regulator 1 |
822 |
0.78 |
| chr17_21226277_21226442 | 0.11 |
MAP2K3 |
mitogen-activated protein kinase kinase 3 |
10905 |
0.2 |
| chr7_19152184_19152434 | 0.11 |
AC003986.6 |
|
212 |
0.92 |
| chr9_130504383_130504534 | 0.11 |
TOR2A |
torsin family 2, member A |
6854 |
0.1 |
| chr9_118918844_118919316 | 0.11 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
2997 |
0.34 |
| chr3_58319217_58319933 | 0.11 |
PXK |
PX domain containing serine/threonine kinase |
542 |
0.78 |
| chr4_186696107_186696537 | 0.11 |
SORBS2 |
sorbin and SH3 domain containing 2 |
108 |
0.98 |
| chr21_26378043_26378194 | 0.11 |
ENSG00000232512 |
. |
95327 |
0.09 |
| chr12_20141265_20141425 | 0.11 |
ENSG00000221679 |
. |
15546 |
0.3 |
| chr13_31481253_31481536 | 0.11 |
MEDAG |
mesenteric estrogen-dependent adipogenesis |
551 |
0.78 |
| chr10_123469524_123469675 | 0.11 |
RP11-78A18.2 |
|
26219 |
0.24 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.2 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0048342 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.2 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.3 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0046398 | UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.0 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.1 | GO:0046174 | polyol catabolic process(GO:0046174) |
| 0.0 | 0.3 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0043032 | positive regulation of macrophage activation(GO:0043032) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0042891 | tetracycline transport(GO:0015904) antibiotic transport(GO:0042891) toxin transport(GO:1901998) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0044091 | membrane biogenesis(GO:0044091) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.0 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.0 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.1 | GO:0006385 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.0 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.0 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.0 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |