Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
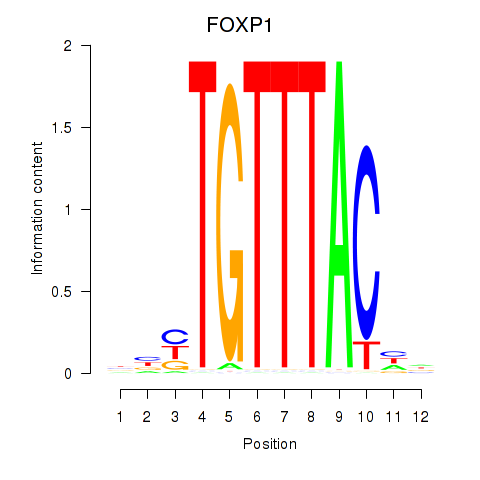
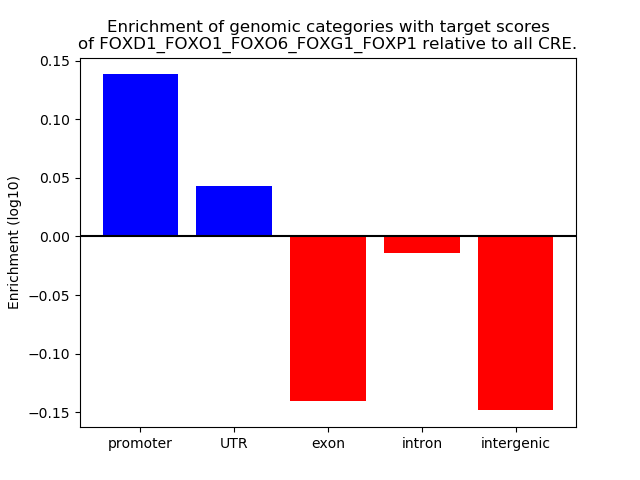
Results for FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
Z-value: 0.48
Transcription factors associated with FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
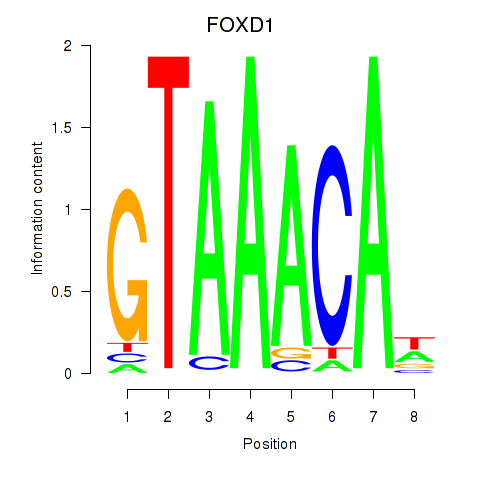
FOXD1
|
ENSG00000251493.2 | forkhead box D1 |
|
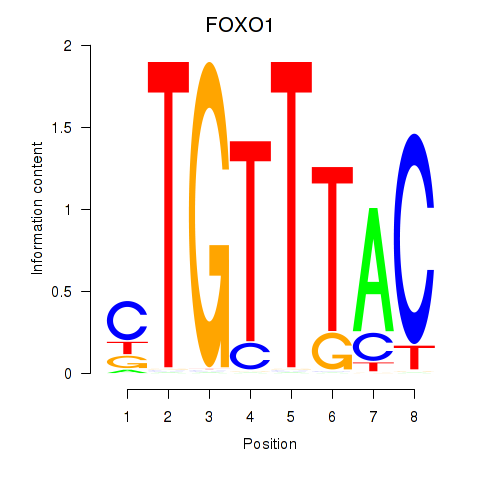
FOXO1
|
ENSG00000150907.6 | forkhead box O1 |
|
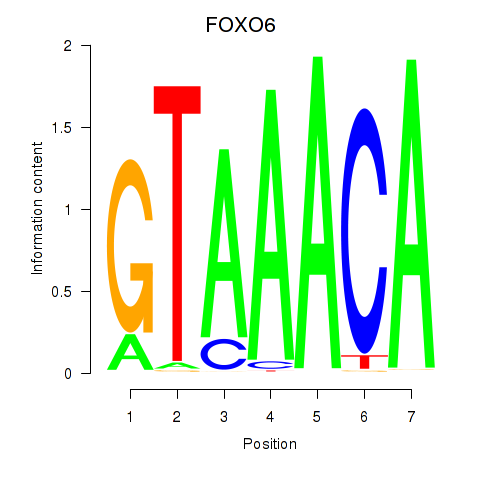
FOXO6
|
ENSG00000204060.4 | forkhead box O6 |
|
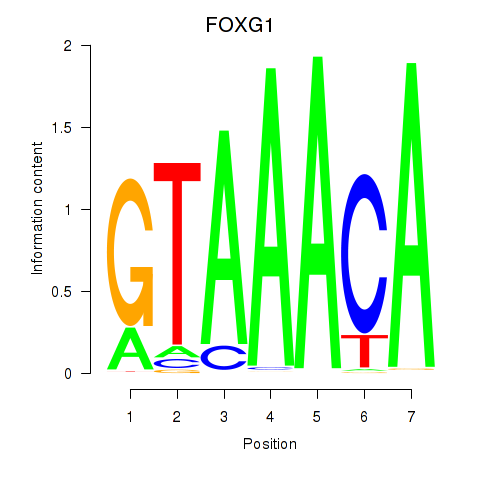
FOXG1
|
ENSG00000176165.7 | forkhead box G1 |
|
FOXP1
|
ENSG00000114861.14 | forkhead box P1 |
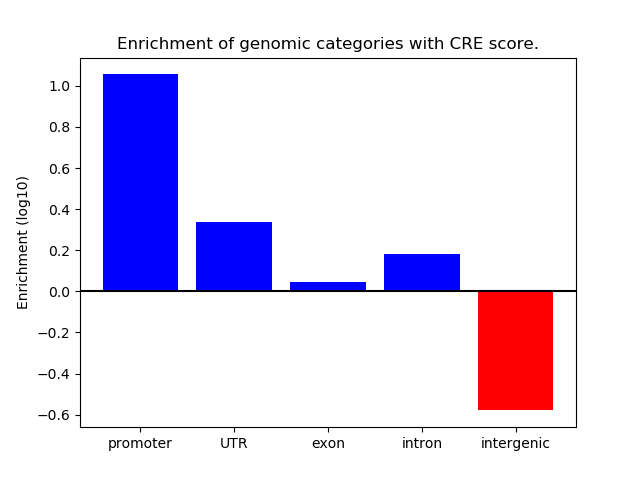
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_72739801_72739952 | FOXD1 | 4476 | 0.237971 | -0.86 | 2.8e-03 | Click! |
| chr5_72746479_72746692 | FOXD1 | 2233 | 0.316554 | -0.82 | 6.6e-03 | Click! |
| chr5_72741608_72741807 | FOXD1 | 2645 | 0.293143 | -0.77 | 1.6e-02 | Click! |
| chr5_72733592_72733937 | FOXD1 | 10588 | 0.210827 | -0.74 | 2.3e-02 | Click! |
| chr5_72661694_72661845 | FOXD1 | 82583 | 0.086668 | -0.71 | 3.1e-02 | Click! |
| chr14_29238229_29238380 | FOXG1 | 2017 | 0.253606 | -0.88 | 1.7e-03 | Click! |
| chr14_29238530_29238681 | FOXG1 | 2318 | 0.227993 | -0.87 | 2.5e-03 | Click! |
| chr14_29235333_29235626 | FOXG1 | 429 | 0.694799 | -0.81 | 7.5e-03 | Click! |
| chr14_29234676_29234957 | FOXG1 | 234 | 0.601383 | -0.77 | 1.4e-02 | Click! |
| chr14_29237083_29237259 | FOXG1 | 884 | 0.510447 | -0.76 | 1.6e-02 | Click! |
| chr13_41227499_41227650 | FOXO1 | 13160 | 0.236041 | 0.83 | 5.9e-03 | Click! |
| chr13_41223921_41224072 | FOXO1 | 16738 | 0.228955 | -0.75 | 2.0e-02 | Click! |
| chr13_41237881_41238133 | FOXO1 | 2727 | 0.331556 | 0.73 | 2.6e-02 | Click! |
| chr13_41219212_41219363 | FOXO1 | 21447 | 0.218996 | -0.70 | 3.4e-02 | Click! |
| chr13_41238416_41238842 | FOXO1 | 2105 | 0.383156 | 0.68 | 4.5e-02 | Click! |
| chr1_41826426_41827003 | FOXO6 | 880 | 0.642751 | 0.57 | 1.1e-01 | Click! |
| chr1_41807666_41807817 | FOXO6 | 19853 | 0.204849 | -0.54 | 1.3e-01 | Click! |
| chr1_41827168_41827846 | FOXO6 | 87 | 0.975968 | 0.42 | 2.6e-01 | Click! |
| chr1_41824027_41824178 | FOXO6 | 3492 | 0.272371 | -0.42 | 2.6e-01 | Click! |
| chr1_41832245_41832396 | FOXO6 | 4726 | 0.239646 | -0.32 | 4.1e-01 | Click! |
| chr3_71150513_71150664 | FOXP1 | 29156 | 0.254969 | -0.86 | 3.0e-03 | Click! |
| chr3_71294181_71294421 | FOXP1 | 15 | 0.986638 | -0.84 | 4.7e-03 | Click! |
| chr3_71350015_71350217 | FOXP1 | 3795 | 0.293781 | 0.83 | 5.4e-03 | Click! |
| chr3_71115551_71115702 | FOXP1 | 708 | 0.812789 | 0.80 | 9.5e-03 | Click! |
| chr3_71157089_71157240 | FOXP1 | 22580 | 0.276377 | -0.79 | 1.1e-02 | Click! |
Activity of the FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 motif across conditions
Conditions sorted by the z-value of the FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
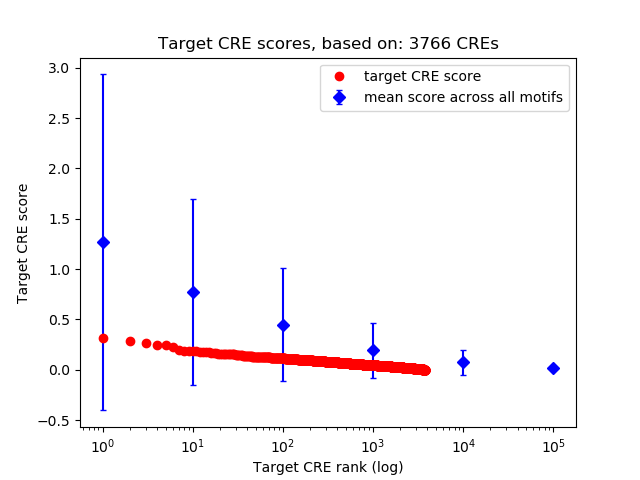
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_89884993_89885181 | 0.31 |
FAM13A |
family with sequence similarity 13, member A |
65964 |
0.13 |
| chr7_150264558_150265605 | 0.28 |
GIMAP4 |
GTPase, IMAP family member 4 |
557 |
0.78 |
| chr3_151917198_151917573 | 0.27 |
MBNL1 |
muscleblind-like splicing regulator 1 |
68444 |
0.12 |
| chr1_198608588_198608957 | 0.25 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
480 |
0.86 |
| chr11_3009712_3009890 | 0.24 |
NAP1L4 |
nucleosome assembly protein 1-like 4 |
588 |
0.57 |
| chr4_154456967_154457118 | 0.23 |
KIAA0922 |
KIAA0922 |
20006 |
0.24 |
| chr6_24951710_24951975 | 0.20 |
FAM65B |
family with sequence similarity 65, member B |
15654 |
0.22 |
| chr2_158298330_158298802 | 0.19 |
ENSG00000216054 |
. |
504 |
0.79 |
| chr6_24934559_24934719 | 0.19 |
FAM65B |
family with sequence similarity 65, member B |
1549 |
0.46 |
| chr2_45794731_45794926 | 0.19 |
SRBD1 |
S1 RNA binding domain 1 |
326 |
0.88 |
| chr6_11727810_11727987 | 0.18 |
ADTRP |
androgen-dependent TFPI-regulating protein |
7991 |
0.25 |
| chrX_70329371_70329522 | 0.18 |
IL2RG |
interleukin 2 receptor, gamma |
328 |
0.79 |
| chr19_17861983_17862176 | 0.17 |
FCHO1 |
FCH domain only 1 |
207 |
0.91 |
| chr5_39218193_39218351 | 0.17 |
FYB |
FYN binding protein |
1393 |
0.56 |
| chr1_84610455_84610717 | 0.17 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
632 |
0.82 |
| chr3_16328270_16328443 | 0.17 |
OXNAD1 |
oxidoreductase NAD-binding domain containing 1 |
17608 |
0.16 |
| chr22_40323366_40323609 | 0.17 |
GRAP2 |
GRB2-related adaptor protein 2 |
846 |
0.6 |
| chr1_209930701_209930998 | 0.17 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
994 |
0.46 |
| chr5_39073301_39073622 | 0.16 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
1030 |
0.63 |
| chr6_33043813_33044241 | 0.16 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
284 |
0.84 |
| chr7_138790255_138790726 | 0.16 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
3610 |
0.26 |
| chr1_12232983_12233134 | 0.16 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
5998 |
0.17 |
| chr3_142663096_142663442 | 0.16 |
RP11-372E1.4 |
|
6187 |
0.16 |
| chr7_5436161_5436613 | 0.16 |
TNRC18 |
trinucleotide repeat containing 18 |
8727 |
0.16 |
| chr16_30484007_30484505 | 0.16 |
ITGAL |
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
184 |
0.81 |
| chrX_70331541_70331692 | 0.16 |
IL2RG |
interleukin 2 receptor, gamma |
135 |
0.92 |
| chr15_91137584_91137808 | 0.15 |
CRTC3 |
CREB regulated transcription coactivator 3 |
751 |
0.57 |
| chr11_44702468_44702619 | 0.15 |
TSPAN18 |
tetraspanin 18 |
45472 |
0.15 |
| chr12_32113291_32114105 | 0.15 |
KIAA1551 |
KIAA1551 |
1345 |
0.51 |
| chr5_39201497_39202174 | 0.15 |
FYB |
FYN binding protein |
1294 |
0.59 |
| chr10_11210404_11210686 | 0.15 |
RP3-323N1.2 |
|
2794 |
0.27 |
| chr8_134071859_134072010 | 0.15 |
SLA |
Src-like-adaptor |
669 |
0.77 |
| chr1_185663060_185663272 | 0.15 |
HMCN1 |
hemicentin 1 |
40517 |
0.17 |
| chr13_41372571_41372761 | 0.14 |
SLC25A15 |
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 |
5372 |
0.18 |
| chr17_76776568_76776719 | 0.14 |
CYTH1 |
cytohesin 1 |
1711 |
0.35 |
| chr10_22944496_22944670 | 0.14 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
58454 |
0.15 |
| chr1_150975096_150975673 | 0.14 |
FAM63A |
family with sequence similarity 63, member A |
3577 |
0.1 |
| chr15_66083115_66083406 | 0.14 |
DENND4A |
DENN/MADD domain containing 4A |
1202 |
0.45 |
| chr11_104971052_104971203 | 0.14 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
1031 |
0.28 |
| chr6_152505186_152505339 | 0.13 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
15763 |
0.29 |
| chr6_53200265_53200465 | 0.13 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
13222 |
0.2 |
| chr21_44104798_44104999 | 0.13 |
PDE9A |
phosphodiesterase 9A |
30975 |
0.15 |
| chr3_45986008_45986159 | 0.13 |
CXCR6 |
chemokine (C-X-C motif) receptor 6 |
428 |
0.8 |
| chr1_198492691_198493057 | 0.13 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
16930 |
0.28 |
| chr6_41906196_41906466 | 0.13 |
CCND3 |
cyclin D3 |
2133 |
0.22 |
| chr19_42946806_42946957 | 0.13 |
CXCL17 |
chemokine (C-X-C motif) ligand 17 |
319 |
0.82 |
| chr7_852914_853138 | 0.13 |
SUN1 |
Sad1 and UNC84 domain containing 1 |
2502 |
0.27 |
| chr19_13906051_13906257 | 0.13 |
ZSWIM4 |
zinc finger, SWIM-type containing 4 |
120 |
0.9 |
| chr13_99953472_99953749 | 0.13 |
GPR183 |
G protein-coupled receptor 183 |
6049 |
0.22 |
| chr10_71853524_71853675 | 0.13 |
AIFM2 |
apoptosis-inducing factor, mitochondrion-associated, 2 |
30259 |
0.15 |
| chr11_96656080_96656231 | 0.13 |
ENSG00000200411 |
. |
448299 |
0.01 |
| chr12_123713862_123714013 | 0.13 |
MPHOSPH9 |
M-phase phosphoprotein 9 |
1056 |
0.41 |
| chr5_66646304_66646752 | 0.13 |
ENSG00000222939 |
. |
113908 |
0.07 |
| chr1_161154335_161154531 | 0.13 |
B4GALT3 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
7146 |
0.07 |
| chr2_20776643_20776868 | 0.13 |
HS1BP3-IT1 |
HS1BP3 intronic transcript 1 (non-protein coding) |
15553 |
0.21 |
| chr17_66433856_66434617 | 0.13 |
WIPI1 |
WD repeat domain, phosphoinositide interacting 1 |
4524 |
0.23 |
| chr8_125575959_125576110 | 0.13 |
MTSS1 |
metastasis suppressor 1 |
1934 |
0.28 |
| chr11_6766181_6766335 | 0.13 |
GVINP1 |
GTPase, very large interferon inducible pseudogene 1 |
23147 |
0.11 |
| chr6_25035340_25035594 | 0.13 |
ENSG00000244618 |
. |
3957 |
0.21 |
| chr1_181003153_181003921 | 0.13 |
MR1 |
major histocompatibility complex, class I-related |
398 |
0.84 |
| chr7_134832844_134833492 | 0.13 |
AC083862.1 |
Uncharacterized protein |
252 |
0.58 |
| chr14_71448816_71448967 | 0.12 |
PCNX |
pecanex homolog (Drosophila) |
30857 |
0.24 |
| chr2_158291882_158292250 | 0.12 |
CYTIP |
cytohesin 1 interacting protein |
3860 |
0.26 |
| chr14_102861114_102861265 | 0.12 |
TECPR2 |
tectonin beta-propeller repeat containing 2 |
31773 |
0.12 |
| chr1_66839032_66839197 | 0.12 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
19049 |
0.27 |
| chr10_73847731_73848399 | 0.12 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
21 |
0.98 |
| chr2_218654553_218654808 | 0.12 |
DIRC3 |
disrupted in renal carcinoma 3 |
33402 |
0.19 |
| chr1_154926867_154927056 | 0.12 |
PBXIP1 |
pre-B-cell leukemia homeobox interacting protein 1 |
1619 |
0.18 |
| chr3_50643497_50643669 | 0.12 |
CISH |
cytokine inducible SH2-containing protein |
5620 |
0.15 |
| chr3_185826921_185827162 | 0.12 |
ETV5 |
ets variant 5 |
140 |
0.97 |
| chr13_99958832_99958994 | 0.12 |
GPR183 |
G protein-coupled receptor 183 |
746 |
0.69 |
| chr1_207996298_207996642 | 0.12 |
ENSG00000203709 |
. |
20602 |
0.22 |
| chr2_46578889_46579334 | 0.12 |
EPAS1 |
endothelial PAS domain protein 1 |
54570 |
0.13 |
| chr4_56814187_56814608 | 0.12 |
CEP135 |
centrosomal protein 135kDa |
640 |
0.74 |
| chr1_174933757_174934016 | 0.12 |
RABGAP1L |
RAB GTPase activating protein 1-like |
19 |
0.97 |
| chr14_89895919_89896980 | 0.12 |
FOXN3-AS1 |
FOXN3 antisense RNA 1 |
12751 |
0.17 |
| chr3_108205436_108205587 | 0.12 |
MYH15 |
myosin, heavy chain 15 |
42658 |
0.16 |
| chr1_6520532_6520815 | 0.12 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
5017 |
0.12 |
| chr5_96268734_96268944 | 0.12 |
LNPEP |
leucyl/cystinyl aminopeptidase |
2329 |
0.27 |
| chr18_60823743_60823894 | 0.12 |
RP11-299P2.1 |
|
5265 |
0.26 |
| chr22_50630477_50630948 | 0.12 |
RP3-402G11.25 |
|
119 |
0.86 |
| chr5_171611170_171611339 | 0.12 |
STK10 |
serine/threonine kinase 10 |
4136 |
0.22 |
| chr1_186291918_186292069 | 0.12 |
ENSG00000202025 |
. |
10933 |
0.17 |
| chr16_68033325_68033555 | 0.12 |
DPEP2 |
dipeptidase 2 |
19 |
0.94 |
| chr1_111215452_111215765 | 0.12 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
2047 |
0.31 |
| chr7_1086655_1086891 | 0.12 |
GPR146 |
G protein-coupled receptor 146 |
2561 |
0.15 |
| chr4_164479735_164479886 | 0.12 |
ENSG00000264535 |
. |
35366 |
0.17 |
| chr5_39200681_39200881 | 0.12 |
FYB |
FYN binding protein |
2348 |
0.41 |
| chr3_151911560_151911983 | 0.12 |
MBNL1 |
muscleblind-like splicing regulator 1 |
74058 |
0.11 |
| chrX_19858890_19859041 | 0.12 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
41096 |
0.19 |
| chr16_79123602_79124194 | 0.12 |
RP11-556H2.3 |
|
49 |
0.98 |
| chr7_4784274_4784732 | 0.12 |
FOXK1 |
forkhead box K1 |
4013 |
0.21 |
| chr14_53593743_53594005 | 0.12 |
DDHD1 |
DDHD domain containing 1 |
22930 |
0.17 |
| chr7_150413706_150414064 | 0.12 |
GIMAP1 |
GTPase, IMAP family member 1 |
240 |
0.91 |
| chr1_150847141_150847524 | 0.12 |
ARNT |
aryl hydrocarbon receptor nuclear translocator |
1723 |
0.26 |
| chr4_40553021_40553619 | 0.12 |
RBM47 |
RNA binding motif protein 47 |
35330 |
0.17 |
| chr19_3687681_3687872 | 0.12 |
PIP5K1C |
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma |
12612 |
0.1 |
| chr10_12390284_12390435 | 0.11 |
CAMK1D |
calcium/calmodulin-dependent protein kinase ID |
1122 |
0.5 |
| chr1_239881158_239881438 | 0.11 |
ENSG00000233355 |
. |
1068 |
0.49 |
| chr14_98100930_98101414 | 0.11 |
ENSG00000240730 |
. |
104662 |
0.08 |
| chr6_150954132_150954283 | 0.11 |
RP11-136K14.2 |
|
8394 |
0.22 |
| chr14_102289544_102289975 | 0.11 |
CTD-2017C7.2 |
|
13101 |
0.14 |
| chr4_84035760_84036001 | 0.11 |
PLAC8 |
placenta-specific 8 |
9 |
0.98 |
| chr6_144665292_144666004 | 0.11 |
UTRN |
utrophin |
411 |
0.88 |
| chr18_60874185_60874445 | 0.11 |
ENSG00000238988 |
. |
12417 |
0.23 |
| chr2_112177530_112177803 | 0.11 |
ENSG00000266139 |
. |
98998 |
0.09 |
| chr3_187803854_187804005 | 0.11 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
67143 |
0.12 |
| chr7_26412308_26412459 | 0.11 |
AC004540.4 |
|
3509 |
0.27 |
| chr17_46505734_46505885 | 0.11 |
SKAP1 |
src kinase associated phosphoprotein 1 |
1743 |
0.29 |
| chr2_430021_430194 | 0.11 |
FAM150B |
family with sequence similarity 150, member B |
141256 |
0.04 |
| chr20_52226068_52226301 | 0.11 |
ZNF217 |
zinc finger protein 217 |
753 |
0.69 |
| chr17_75103441_75103592 | 0.11 |
ENSG00000234912 |
. |
18127 |
0.18 |
| chr20_55975682_55975833 | 0.11 |
RP4-800J21.3 |
|
7639 |
0.16 |
| chr1_174934321_174935265 | 0.11 |
RABGAP1L |
RAB GTPase activating protein 1-like |
888 |
0.52 |
| chr1_66806954_66807105 | 0.11 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
9157 |
0.31 |
| chr15_34626880_34627184 | 0.11 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
2013 |
0.2 |
| chr14_22631434_22631696 | 0.11 |
ENSG00000238634 |
. |
20678 |
0.25 |
| chr1_25656822_25656973 | 0.11 |
TMEM50A |
transmembrane protein 50A |
7511 |
0.13 |
| chr2_149403004_149403363 | 0.11 |
EPC2 |
enhancer of polycomb homolog 2 (Drosophila) |
194 |
0.97 |
| chr12_113345256_113345451 | 0.11 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
496 |
0.81 |
| chr1_235632222_235632373 | 0.11 |
B3GALNT2 |
beta-1,3-N-acetylgalactosaminyltransferase 2 |
35425 |
0.16 |
| chr1_226068766_226068917 | 0.11 |
TMEM63A |
transmembrane protein 63A |
1228 |
0.35 |
| chr1_226862428_226862635 | 0.11 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
237 |
0.93 |
| chr11_108535331_108535645 | 0.11 |
DDX10 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
264 |
0.95 |
| chr9_117690832_117691467 | 0.11 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
1548 |
0.52 |
| chr2_87830138_87830289 | 0.11 |
RP11-1399P15.1 |
|
52660 |
0.16 |
| chr2_198170219_198170545 | 0.11 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
3139 |
0.21 |
| chr7_44665248_44665399 | 0.11 |
OGDH |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
1262 |
0.46 |
| chr11_102192518_102192994 | 0.11 |
BIRC3 |
baculoviral IAP repeat containing 3 |
3196 |
0.22 |
| chr6_159465648_159465970 | 0.11 |
TAGAP |
T-cell activation RhoGTPase activating protein |
241 |
0.93 |
| chr10_17687713_17687972 | 0.11 |
STAM |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
1578 |
0.29 |
| chr12_122239467_122240363 | 0.11 |
RHOF |
ras homolog family member F (in filopodia) |
919 |
0.49 |
| chr6_25138653_25139026 | 0.11 |
ENSG00000222373 |
. |
53802 |
0.11 |
| chr9_88953252_88953465 | 0.11 |
ZCCHC6 |
zinc finger, CCHC domain containing 6 |
4680 |
0.26 |
| chr13_74806186_74806399 | 0.11 |
ENSG00000206617 |
. |
57059 |
0.16 |
| chr19_16493261_16493427 | 0.11 |
ENSG00000243745 |
. |
16426 |
0.13 |
| chr1_172149860_172150165 | 0.11 |
DNM3OS |
DNM3 opposite strand/antisense RNA |
36078 |
0.15 |
| chr2_161995820_161996335 | 0.10 |
TANK |
TRAF family member-associated NFKB activator |
2611 |
0.33 |
| chr6_10528716_10529334 | 0.10 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
436 |
0.83 |
| chr2_220113848_220114166 | 0.10 |
STK16 |
serine/threonine kinase 16 |
3389 |
0.09 |
| chr2_68430764_68431023 | 0.10 |
RP11-474G23.2 |
|
23928 |
0.12 |
| chr4_80979447_80979859 | 0.10 |
ANTXR2 |
anthrax toxin receptor 2 |
14064 |
0.28 |
| chr5_1800151_1800360 | 0.10 |
MRPL36 |
mitochondrial ribosomal protein L36 |
246 |
0.88 |
| chr3_111266223_111266431 | 0.10 |
CD96 |
CD96 molecule |
5330 |
0.28 |
| chr1_160547631_160547822 | 0.10 |
CD84 |
CD84 molecule |
1537 |
0.33 |
| chr17_65241820_65242054 | 0.10 |
HELZ |
helicase with zinc finger |
168 |
0.89 |
| chr12_6744541_6744794 | 0.10 |
LPAR5 |
lysophosphatidic acid receptor 5 |
946 |
0.33 |
| chr9_129286057_129286218 | 0.10 |
ENSG00000221768 |
. |
7133 |
0.22 |
| chr13_50203416_50203569 | 0.10 |
ARL11 |
ADP-ribosylation factor-like 11 |
1057 |
0.56 |
| chr1_8455637_8455851 | 0.10 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
27982 |
0.15 |
| chrX_72800965_72801116 | 0.10 |
CHIC1 |
cysteine-rich hydrophobic domain 1 |
17996 |
0.28 |
| chr4_16226606_16226757 | 0.10 |
TAPT1 |
transmembrane anterior posterior transformation 1 |
1413 |
0.36 |
| chr1_111417492_111418003 | 0.10 |
CD53 |
CD53 molecule |
1971 |
0.34 |
| chr5_134240090_134240263 | 0.10 |
PCBD2 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
420 |
0.81 |
| chr10_17726067_17726351 | 0.10 |
ENSG00000251959 |
. |
5022 |
0.18 |
| chr20_47436634_47436796 | 0.10 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
7705 |
0.28 |
| chr7_134832355_134832754 | 0.10 |
TMEM140 |
transmembrane protein 140 |
270 |
0.58 |
| chr11_75063940_75064091 | 0.10 |
ARRB1 |
arrestin, beta 1 |
1142 |
0.41 |
| chr10_21914636_21914787 | 0.10 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
25972 |
0.17 |
| chr21_28334749_28335004 | 0.10 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
3956 |
0.26 |
| chrX_24498316_24498580 | 0.10 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
14875 |
0.2 |
| chr19_6086785_6086936 | 0.10 |
CTC-232P5.3 |
|
18896 |
0.13 |
| chr14_22920576_22920727 | 0.10 |
TRDD3 |
T cell receptor delta diversity 3 |
2546 |
0.16 |
| chr15_81594027_81594306 | 0.10 |
IL16 |
interleukin 16 |
2409 |
0.29 |
| chr22_24540304_24540599 | 0.10 |
CABIN1 |
calcineurin binding protein 1 |
11355 |
0.17 |
| chr19_2428027_2428178 | 0.10 |
TIMM13 |
translocase of inner mitochondrial membrane 13 homolog (yeast) |
210 |
0.91 |
| chr11_45069841_45069992 | 0.10 |
PRDM11 |
PR domain containing 11 |
45648 |
0.17 |
| chr7_157145359_157145611 | 0.10 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
12970 |
0.24 |
| chr5_140997567_140997853 | 0.10 |
AC008781.7 |
|
271 |
0.75 |
| chr7_129360327_129360478 | 0.10 |
ENSG00000212238 |
. |
35704 |
0.13 |
| chr15_86233199_86233602 | 0.10 |
RP11-815J21.1 |
|
10940 |
0.11 |
| chr1_70875957_70876778 | 0.10 |
CTH |
cystathionase (cystathionine gamma-lyase) |
534 |
0.81 |
| chr7_1572516_1572667 | 0.10 |
MAFK |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
2241 |
0.23 |
| chr4_103425480_103425657 | 0.10 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
1036 |
0.55 |
| chr11_6767215_6767876 | 0.10 |
OR2AG2 |
olfactory receptor, family 2, subfamily AG, member 2 |
22741 |
0.11 |
| chr13_41554167_41554365 | 0.10 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
2152 |
0.33 |
| chr1_241715191_241715459 | 0.10 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
16610 |
0.2 |
| chr6_127045549_127045700 | 0.10 |
ENSG00000201613 |
. |
134085 |
0.05 |
| chr8_126442398_126442549 | 0.10 |
TRIB1 |
tribbles pseudokinase 1 |
90 |
0.98 |
| chr1_100573281_100573432 | 0.10 |
RP4-714D9.2 |
|
23329 |
0.12 |
| chr3_128838581_128838923 | 0.10 |
RAB43 |
RAB43, member RAS oncogene family |
1872 |
0.23 |
| chrX_3729988_3730150 | 0.09 |
RP11-706O15.1 |
HCG1981372, isoform CRA_c; Uncharacterized protein |
31829 |
0.22 |
| chr12_102222797_102222948 | 0.09 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
1585 |
0.27 |
| chr2_201987013_201987300 | 0.09 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
44 |
0.96 |
| chr19_39894109_39894559 | 0.09 |
ZFP36 |
ZFP36 ring finger protein |
3119 |
0.1 |
| chr13_41554727_41554878 | 0.09 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
1616 |
0.4 |
| chr8_10367956_10368344 | 0.09 |
PRSS51 |
protease, serine, 51 |
7375 |
0.19 |
| chr21_46337622_46337773 | 0.09 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
3073 |
0.14 |
| chr11_119136952_119137103 | 0.09 |
ENSG00000222249 |
. |
23360 |
0.1 |
| chr1_107932269_107932420 | 0.09 |
ENSG00000238883 |
. |
58833 |
0.16 |
| chr11_62476686_62476935 | 0.09 |
BSCL2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
18 |
0.92 |
| chr9_138967584_138968045 | 0.09 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
19317 |
0.19 |
| chr2_71644611_71645120 | 0.09 |
ZNF638 |
zinc finger protein 638 |
10496 |
0.21 |
| chrX_44748789_44748940 | 0.09 |
ENSG00000252113 |
. |
4113 |
0.25 |
| chr15_71740308_71740459 | 0.09 |
ENSG00000252813 |
. |
27443 |
0.23 |
| chr13_41540610_41540946 | 0.09 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
15640 |
0.18 |
| chr12_4937651_4937802 | 0.09 |
RP3-377H17.2 |
|
10395 |
0.2 |
| chr12_110517460_110517751 | 0.09 |
C12orf76 |
chromosome 12 open reading frame 76 |
6114 |
0.22 |
| chr3_52288072_52288223 | 0.09 |
PPM1M |
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
5170 |
0.1 |
| chrX_40028928_40029079 | 0.09 |
BCOR |
BCL6 corepressor |
7570 |
0.32 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0019348 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.1 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.1 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.0 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0061526 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0075713 | establishment of viral latency(GO:0019043) establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.2 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.0 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.0 | GO:0033088 | regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.0 | GO:0061047 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0032142 | single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.0 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.0 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |