Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
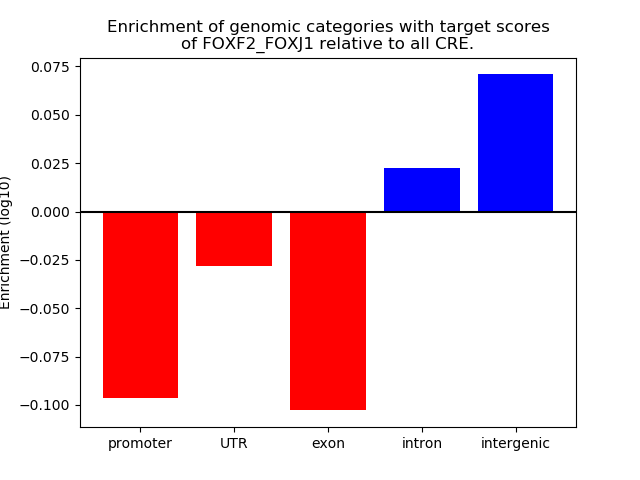
Results for FOXF2_FOXJ1
Z-value: 0.84
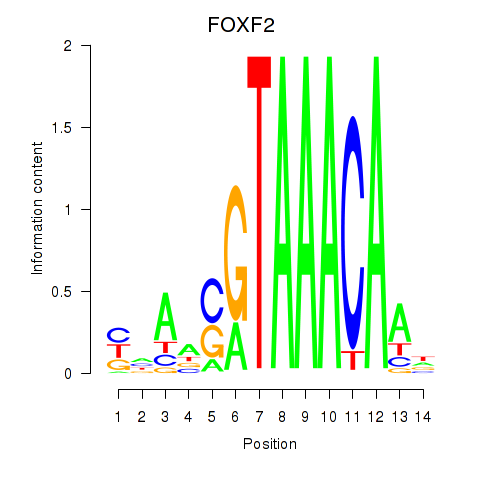
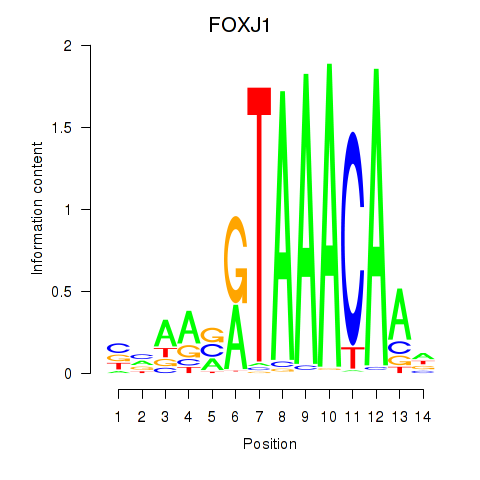
Transcription factors associated with FOXF2_FOXJ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXF2
|
ENSG00000137273.3 | forkhead box F2 |
|
FOXJ1
|
ENSG00000129654.7 | forkhead box J1 |
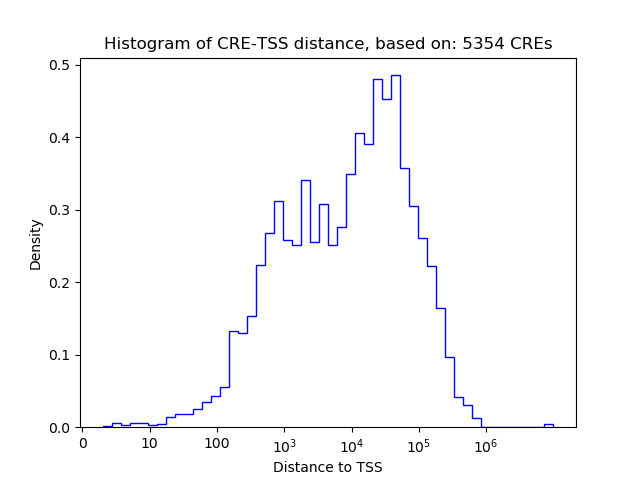
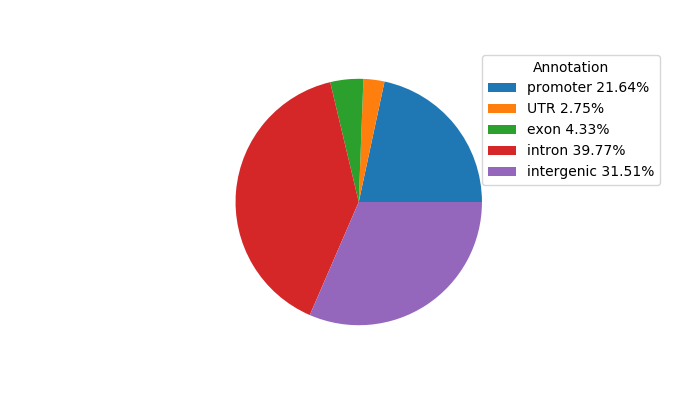
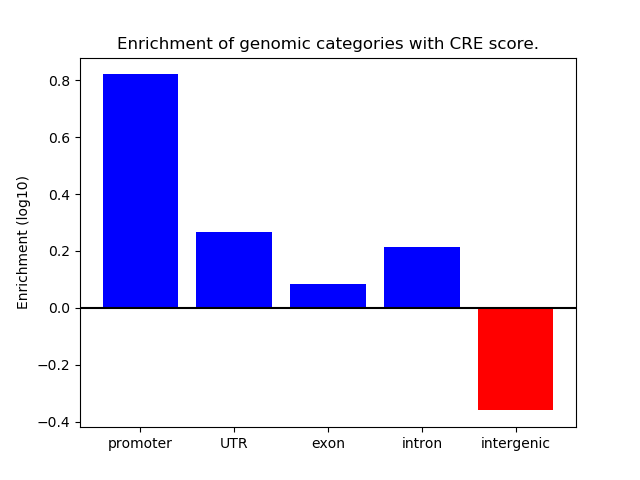
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_1393770_1393965 | FOXF2 | 3798 | 0.264951 | -0.52 | 1.6e-01 | Click! |
| chr6_1389874_1390147 | FOXF2 | 59 | 0.978586 | 0.36 | 3.4e-01 | Click! |
| chr6_1391347_1391718 | FOXF2 | 1463 | 0.453743 | 0.29 | 4.5e-01 | Click! |
| chr6_1392834_1392985 | FOXF2 | 2840 | 0.298522 | -0.27 | 4.8e-01 | Click! |
| chr6_1407500_1407651 | FOXF2 | 17506 | 0.209936 | 0.27 | 4.9e-01 | Click! |
| chr17_74137094_74137375 | FOXJ1 | 146 | 0.736402 | -0.37 | 3.3e-01 | Click! |
Activity of the FOXF2_FOXJ1 motif across conditions
Conditions sorted by the z-value of the FOXF2_FOXJ1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
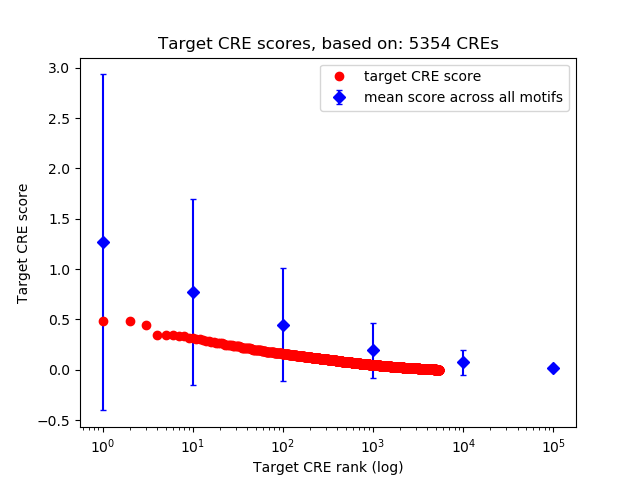
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_120466546_120467060 | 0.49 |
TLR4 |
toll-like receptor 4 |
153 |
0.96 |
| chr8_15398472_15398721 | 0.49 |
TUSC3 |
tumor suppressor candidate 3 |
804 |
0.72 |
| chr4_71587677_71588145 | 0.44 |
RUFY3 |
RUN and FYVE domain containing 3 |
183 |
0.92 |
| chr8_107738861_107739281 | 0.35 |
OXR1 |
oxidation resistance 1 |
658 |
0.71 |
| chr1_109913452_109914259 | 0.34 |
SORT1 |
sortilin 1 |
22124 |
0.14 |
| chr5_59893443_59893927 | 0.34 |
DEPDC1B |
DEP domain containing 1B |
102276 |
0.07 |
| chr7_19152184_19152434 | 0.34 |
AC003986.6 |
|
212 |
0.92 |
| chr2_152214605_152215087 | 0.33 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
740 |
0.63 |
| chr10_13569706_13570278 | 0.32 |
RP11-214D15.2 |
|
524 |
0.45 |
| chr7_79449658_79449850 | 0.32 |
ENSG00000206818 |
. |
91770 |
0.09 |
| chr2_27995563_27995714 | 0.30 |
MRPL33 |
mitochondrial ribosomal protein L33 |
1054 |
0.48 |
| chr6_151648337_151648657 | 0.30 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
1674 |
0.27 |
| chr9_128235766_128235975 | 0.30 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
10910 |
0.23 |
| chr6_44510355_44510599 | 0.29 |
ENSG00000266619 |
. |
107099 |
0.06 |
| chr1_216773992_216774229 | 0.28 |
ESRRG |
estrogen-related receptor gamma |
122564 |
0.06 |
| chr4_115282272_115282768 | 0.28 |
UGT8 |
UDP glycosyltransferase 8 |
237091 |
0.02 |
| chr6_132268442_132268593 | 0.28 |
RP11-69I8.3 |
|
3569 |
0.25 |
| chr6_136223158_136223344 | 0.27 |
ENSG00000238921 |
. |
12642 |
0.24 |
| chr6_153269357_153269511 | 0.27 |
FBXO5 |
F-box protein 5 |
34719 |
0.14 |
| chr8_48571824_48572102 | 0.27 |
SPIDR |
scaffolding protein involved in DNA repair |
263 |
0.93 |
| chr3_59803963_59804385 | 0.27 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
153409 |
0.05 |
| chr7_19308580_19308778 | 0.25 |
FERD3L |
Fer3-like bHLH transcription factor |
123635 |
0.05 |
| chr14_79747812_79748200 | 0.25 |
NRXN3 |
neurexin 3 |
1757 |
0.44 |
| chr13_37493387_37493814 | 0.25 |
SMAD9 |
SMAD family member 9 |
775 |
0.69 |
| chr18_25756118_25756718 | 0.24 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
992 |
0.73 |
| chr11_36616853_36617082 | 0.24 |
C11orf74 |
chromosome 11 open reading frame 74 |
567 |
0.5 |
| chr19_54534469_54534642 | 0.24 |
VSTM1 |
V-set and transmembrane domain containing 1 |
27122 |
0.08 |
| chr14_52707563_52708006 | 0.24 |
PTGDR |
prostaglandin D2 receptor (DP) |
26647 |
0.22 |
| chr18_65983753_65984295 | 0.24 |
TMX3 |
thioredoxin-related transmembrane protein 3 |
398270 |
0.01 |
| chr1_172110530_172111247 | 0.23 |
ENSG00000207949 |
. |
2841 |
0.22 |
| chr10_45473981_45474319 | 0.23 |
C10orf10 |
chromosome 10 open reading frame 10 |
87 |
0.95 |
| chr12_21981553_21981788 | 0.23 |
RP11-729I10.2 |
|
1526 |
0.43 |
| chr9_75486718_75486907 | 0.23 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
81159 |
0.11 |
| chr1_60031328_60031726 | 0.22 |
FGGY |
FGGY carbohydrate kinase domain containing |
11937 |
0.3 |
| chr5_90515283_90515434 | 0.22 |
ENSG00000199643 |
. |
51186 |
0.18 |
| chr21_17442999_17443232 | 0.22 |
ENSG00000252273 |
. |
35286 |
0.24 |
| chr12_19392736_19392931 | 0.22 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
2975 |
0.32 |
| chr7_148232370_148232768 | 0.22 |
C7orf33 |
chromosome 7 open reading frame 33 |
55088 |
0.14 |
| chr13_33803329_33803516 | 0.22 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
23279 |
0.2 |
| chr2_45200338_45200489 | 0.21 |
SIX3-AS1 |
SIX3 antisense RNA 1 |
31401 |
0.17 |
| chr10_49734527_49734828 | 0.21 |
ARHGAP22 |
Rho GTPase activating protein 22 |
2396 |
0.33 |
| chr3_87039220_87039752 | 0.21 |
VGLL3 |
vestigial like 3 (Drosophila) |
366 |
0.93 |
| chr16_88628658_88628817 | 0.21 |
ZC3H18 |
zinc finger CCCH-type containing 18 |
8052 |
0.16 |
| chr3_190530283_190530682 | 0.21 |
GMNC |
geminin coiled-coil domain containing |
49922 |
0.18 |
| chr2_185461919_185462338 | 0.21 |
ZNF804A |
zinc finger protein 804A |
965 |
0.73 |
| chr12_1583558_1583788 | 0.21 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
55384 |
0.12 |
| chr1_182931948_182932121 | 0.20 |
ENSG00000264768 |
. |
3324 |
0.2 |
| chr14_59215785_59216332 | 0.20 |
ENSG00000221427 |
. |
21906 |
0.26 |
| chr3_18153147_18153551 | 0.20 |
RP11-158G18.1 |
|
297778 |
0.01 |
| chr22_36367515_36367713 | 0.20 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
9900 |
0.29 |
| chr16_25047588_25047739 | 0.20 |
ARHGAP17 |
Rho GTPase activating protein 17 |
20676 |
0.23 |
| chr5_141980216_141980760 | 0.20 |
AC005592.2 |
|
19075 |
0.18 |
| chr2_74104795_74104946 | 0.20 |
ACTG2 |
actin, gamma 2, smooth muscle, enteric |
15224 |
0.17 |
| chr15_61044545_61044696 | 0.20 |
RP11-554D20.1 |
|
11485 |
0.22 |
| chr8_38855409_38855789 | 0.19 |
ADAM9 |
ADAM metallopeptidase domain 9 |
1094 |
0.34 |
| chr7_27217405_27217623 | 0.19 |
RP1-170O19.20 |
Uncharacterized protein |
2118 |
0.1 |
| chr15_99864758_99865035 | 0.19 |
AC022819.2 |
Uncharacterized protein |
5068 |
0.23 |
| chr2_38008128_38008279 | 0.19 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
42592 |
0.19 |
| chr5_131893791_131893942 | 0.19 |
RAD50 |
RAD50 homolog (S. cerevisiae) |
943 |
0.42 |
| chr12_15038397_15038786 | 0.19 |
MGP |
matrix Gla protein |
200 |
0.92 |
| chr15_85590935_85591086 | 0.19 |
PDE8A |
phosphodiesterase 8A |
65798 |
0.1 |
| chr10_126843486_126843769 | 0.18 |
CTBP2 |
C-terminal binding protein 2 |
3658 |
0.35 |
| chr8_48297219_48297488 | 0.18 |
SPIDR |
scaffolding protein involved in DNA repair |
55610 |
0.16 |
| chr18_52990549_52990700 | 0.18 |
TCF4 |
transcription factor 4 |
1099 |
0.65 |
| chr13_31481253_31481536 | 0.18 |
MEDAG |
mesenteric estrogen-dependent adipogenesis |
551 |
0.78 |
| chr21_17584209_17584483 | 0.18 |
ENSG00000201025 |
. |
72743 |
0.13 |
| chr2_176992012_176992333 | 0.18 |
HOXD8 |
homeobox D8 |
2250 |
0.12 |
| chr10_134389068_134389378 | 0.18 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
32207 |
0.18 |
| chr1_198859885_198860036 | 0.18 |
ENSG00000207759 |
. |
31678 |
0.2 |
| chr3_119921742_119921893 | 0.18 |
ENSG00000206721 |
. |
7350 |
0.22 |
| chr20_9048816_9049419 | 0.18 |
PLCB4 |
phospholipase C, beta 4 |
293 |
0.9 |
| chr7_127293840_127294146 | 0.18 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
1759 |
0.28 |
| chr21_28337250_28337401 | 0.18 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
1507 |
0.46 |
| chr5_123493061_123493232 | 0.18 |
CSNK1G3 |
casein kinase 1, gamma 3 |
569027 |
0.0 |
| chr2_146281977_146282128 | 0.17 |
ENSG00000253036 |
. |
189414 |
0.03 |
| chr1_65730855_65731313 | 0.17 |
DNAJC6 |
DnaJ (Hsp40) homolog, subfamily C, member 6 |
647 |
0.77 |
| chr2_63971666_63971817 | 0.17 |
ENSG00000221085 |
. |
49072 |
0.16 |
| chr3_114477226_114477377 | 0.17 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
486 |
0.86 |
| chr20_1103594_1103745 | 0.17 |
PSMF1 |
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
4389 |
0.25 |
| chr2_43406321_43406536 | 0.17 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
47320 |
0.15 |
| chr3_186539235_186539386 | 0.17 |
RP11-573D15.3 |
|
13829 |
0.08 |
| chr22_46988879_46989444 | 0.17 |
GRAMD4 |
GRAM domain containing 4 |
16135 |
0.21 |
| chr5_67323714_67323865 | 0.17 |
ENSG00000223149 |
. |
60516 |
0.15 |
| chr3_9389156_9389527 | 0.17 |
THUMPD3 |
THUMP domain containing 3 |
15185 |
0.15 |
| chr3_73769050_73769270 | 0.17 |
PDZRN3 |
PDZ domain containing ring finger 3 |
95069 |
0.08 |
| chr6_113970392_113970641 | 0.17 |
ENSG00000266650 |
. |
46399 |
0.16 |
| chr12_30947588_30947773 | 0.17 |
CAPRIN2 |
caprin family member 2 |
39795 |
0.19 |
| chr11_57529774_57530544 | 0.17 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
869 |
0.5 |
| chr8_42394798_42395191 | 0.17 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
1203 |
0.3 |
| chr13_73712912_73713063 | 0.16 |
ENSG00000265959 |
. |
14040 |
0.19 |
| chr9_110400851_110401256 | 0.16 |
ENSG00000202308 |
. |
24474 |
0.24 |
| chr13_21790667_21790818 | 0.16 |
MRP63 |
mitochondrial ribosomal protein 63 |
39958 |
0.12 |
| chr16_67512921_67513072 | 0.16 |
ATP6V0D1 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
1986 |
0.17 |
| chr12_76144582_76144847 | 0.16 |
ENSG00000264062 |
. |
26393 |
0.22 |
| chr3_15847185_15847355 | 0.16 |
ANKRD28 |
ankyrin repeat domain 28 |
9242 |
0.22 |
| chrX_22318596_22318928 | 0.16 |
ZNF645 |
zinc finger protein 645 |
27697 |
0.24 |
| chr2_181056389_181056548 | 0.16 |
CWC22 |
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
184628 |
0.03 |
| chr4_123795073_123795451 | 0.16 |
RP11-170N16.3 |
|
10605 |
0.18 |
| chr1_98369641_98369792 | 0.16 |
DPYD |
dihydropyrimidine dehydrogenase |
16837 |
0.28 |
| chr6_21666737_21667013 | 0.16 |
SOX4 |
SRY (sex determining region Y)-box 4 |
72804 |
0.14 |
| chr7_3632163_3632314 | 0.16 |
AC011284.3 |
|
51178 |
0.18 |
| chr18_57576623_57576774 | 0.16 |
PMAIP1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
9450 |
0.26 |
| chr6_169471024_169471293 | 0.16 |
XXyac-YX65C7_A.2 |
|
142191 |
0.05 |
| chr2_222761989_222762140 | 0.16 |
EPHA4 |
EPH receptor A4 |
323142 |
0.01 |
| chr15_68155525_68155920 | 0.16 |
ENSG00000206625 |
. |
23339 |
0.22 |
| chr8_2094260_2094411 | 0.15 |
MYOM2 |
myomesin 2 |
101151 |
0.08 |
| chr19_11305569_11305933 | 0.15 |
KANK2 |
KN motif and ankyrin repeat domains 2 |
117 |
0.94 |
| chr12_32688080_32688676 | 0.15 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
1120 |
0.61 |
| chr2_167231853_167232143 | 0.15 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
499 |
0.84 |
| chr13_47022181_47022332 | 0.15 |
KIAA0226L |
KIAA0226-like |
9931 |
0.22 |
| chr10_4541408_4541559 | 0.15 |
ENSG00000207124 |
. |
15661 |
0.3 |
| chr1_183512654_183512805 | 0.15 |
NCF2 |
neutrophil cytosolic factor 2 |
25631 |
0.17 |
| chr13_115030758_115030909 | 0.15 |
ENSG00000265450 |
. |
8470 |
0.15 |
| chr10_131755852_131756003 | 0.15 |
EBF3 |
early B-cell factor 3 |
6178 |
0.32 |
| chr2_120189452_120190154 | 0.15 |
TMEM37 |
transmembrane protein 37 |
358 |
0.88 |
| chr12_8090559_8091101 | 0.15 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
1959 |
0.27 |
| chr2_240971693_240971844 | 0.15 |
OR6B2 |
olfactory receptor, family 6, subfamily B, member 2 |
1862 |
0.27 |
| chr6_56573305_56573456 | 0.15 |
DST |
dystonin |
65586 |
0.13 |
| chr5_39424296_39424549 | 0.15 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
548 |
0.85 |
| chr4_187901309_187901460 | 0.15 |
ENSG00000252382 |
. |
122774 |
0.06 |
| chr8_67088876_67089283 | 0.15 |
CRH |
corticotropin releasing hormone |
1881 |
0.44 |
| chr20_47942346_47942497 | 0.15 |
ENSG00000212304 |
. |
45201 |
0.1 |
| chr8_17353947_17354148 | 0.15 |
SLC7A2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
550 |
0.83 |
| chr12_13351250_13351500 | 0.15 |
EMP1 |
epithelial membrane protein 1 |
1655 |
0.47 |
| chr1_120674474_120674625 | 0.15 |
ENSG00000207149 |
. |
4987 |
0.29 |
| chr3_105084192_105084343 | 0.15 |
ALCAM |
activated leukocyte cell adhesion molecule |
1486 |
0.6 |
| chr4_169554219_169554371 | 0.15 |
PALLD |
palladin, cytoskeletal associated protein |
1527 |
0.43 |
| chr9_13719940_13720091 | 0.15 |
MPDZ |
multiple PDZ domain protein |
440426 |
0.01 |
| chr1_158969050_158969253 | 0.15 |
IFI16 |
interferon, gamma-inducible protein 16 |
607 |
0.76 |
| chr2_198539361_198539512 | 0.15 |
RFTN2 |
raftlin family member 2 |
1283 |
0.45 |
| chr17_31255277_31255844 | 0.15 |
TMEM98 |
transmembrane protein 98 |
332 |
0.89 |
| chr3_112013106_112013257 | 0.15 |
SLC9C1 |
solute carrier family 9, subfamily C (Na+-transporting carboxylic acid decarboxylase), member 1 |
76 |
0.98 |
| chr9_16654239_16654390 | 0.15 |
RP11-62F24.1 |
|
28640 |
0.21 |
| chr17_70443074_70443225 | 0.15 |
ENSG00000200783 |
. |
217142 |
0.02 |
| chr10_44304922_44305073 | 0.15 |
ZNF32 |
zinc finger protein 32 |
160693 |
0.03 |
| chr10_28967900_28968168 | 0.15 |
BAMBI |
BMP and activin membrane-bound inhibitor |
1763 |
0.35 |
| chr18_9017410_9017607 | 0.15 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
85120 |
0.08 |
| chr4_74904304_74905084 | 0.14 |
CXCL3 |
chemokine (C-X-C motif) ligand 3 |
288 |
0.87 |
| chr7_22603039_22603190 | 0.14 |
AC002480.3 |
|
158 |
0.96 |
| chr5_158476960_158477151 | 0.14 |
EBF1 |
early B-cell factor 1 |
49646 |
0.14 |
| chr16_86705742_86705893 | 0.14 |
FOXL1 |
forkhead box L1 |
93702 |
0.08 |
| chr9_107731134_107731381 | 0.14 |
ABCA1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
40739 |
0.17 |
| chr13_109567770_109568133 | 0.14 |
MYO16 |
myosin XVI |
29434 |
0.26 |
| chr20_52539385_52539536 | 0.14 |
AC005220.3 |
|
17239 |
0.25 |
| chr12_104199013_104199284 | 0.14 |
RP11-650K20.2 |
|
14240 |
0.14 |
| chr21_17569279_17569619 | 0.14 |
ENSG00000201025 |
. |
87640 |
0.1 |
| chr18_475431_475606 | 0.14 |
COLEC12 |
collectin sub-family member 12 |
25204 |
0.17 |
| chr10_31353638_31353789 | 0.14 |
ENSG00000263578 |
. |
1263 |
0.56 |
| chr7_120664875_120665040 | 0.14 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
35281 |
0.16 |
| chr9_93558631_93558871 | 0.14 |
SYK |
spleen tyrosine kinase |
5318 |
0.35 |
| chr4_109542934_109543085 | 0.14 |
RPL34 |
ribosomal protein L34 |
1197 |
0.48 |
| chr12_81472525_81472676 | 0.14 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
764 |
0.72 |
| chr4_129557030_129557181 | 0.14 |
ENSG00000238802 |
. |
120076 |
0.06 |
| chr17_13503336_13503489 | 0.14 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
1832 |
0.45 |
| chr15_56756676_56757268 | 0.14 |
MNS1 |
meiosis-specific nuclear structural 1 |
363 |
0.92 |
| chr5_92333922_92334106 | 0.14 |
ENSG00000221810 |
. |
280794 |
0.01 |
| chr18_66499529_66499680 | 0.14 |
CCDC102B |
coiled-coil domain containing 102B |
4397 |
0.25 |
| chr2_134325837_134326056 | 0.14 |
NCKAP5 |
NCK-associated protein 5 |
85 |
0.98 |
| chr15_28423630_28423781 | 0.14 |
HERC2 |
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
4136 |
0.31 |
| chr5_74634021_74634424 | 0.14 |
HMGCR |
3-hydroxy-3-methylglutaryl-CoA reductase |
264 |
0.89 |
| chr4_81188224_81188744 | 0.14 |
FGF5 |
fibroblast growth factor 5 |
691 |
0.76 |
| chr8_97468793_97469144 | 0.14 |
ENSG00000202095 |
. |
25580 |
0.2 |
| chr1_246750885_246751091 | 0.14 |
RP11-439E19.1 |
|
18187 |
0.16 |
| chr18_60774564_60774715 | 0.14 |
RP11-299P2.1 |
|
43914 |
0.17 |
| chr7_66532196_66532424 | 0.13 |
ENSG00000266525 |
. |
47074 |
0.12 |
| chr13_111856007_111856158 | 0.13 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
667 |
0.76 |
| chr16_73091234_73091750 | 0.13 |
ZFHX3 |
zinc finger homeobox 3 |
2105 |
0.37 |
| chr11_16813436_16813790 | 0.13 |
PLEKHA7 |
pleckstrin homology domain containing, family A member 7 |
20417 |
0.2 |
| chr7_150485274_150485425 | 0.13 |
TMEM176B |
transmembrane protein 176B |
8471 |
0.16 |
| chr1_183791724_183791999 | 0.13 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
17571 |
0.24 |
| chr7_136831692_136831856 | 0.13 |
hsa-mir-490 |
hsa-mir-490 |
17243 |
0.28 |
| chr4_177711668_177712016 | 0.13 |
VEGFC |
vascular endothelial growth factor C |
2039 |
0.48 |
| chr6_150177526_150177677 | 0.13 |
RP11-350J20.12 |
|
4003 |
0.14 |
| chr18_28681249_28681463 | 0.13 |
RP11-408H20.2 |
|
195 |
0.81 |
| chr16_65152808_65153250 | 0.13 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
2804 |
0.44 |
| chr8_116850745_116850896 | 0.13 |
TRPS1 |
trichorhinophalangeal syndrome I |
28921 |
0.26 |
| chr19_10927638_10927909 | 0.13 |
ENSG00000207752 |
. |
399 |
0.76 |
| chr4_189897651_189897802 | 0.13 |
ENSG00000252275 |
. |
259847 |
0.02 |
| chr3_81873946_81874241 | 0.13 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
62781 |
0.16 |
| chr3_151976533_151976804 | 0.13 |
MBNL1 |
muscleblind-like splicing regulator 1 |
9161 |
0.23 |
| chr5_39547598_39547749 | 0.13 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
85271 |
0.11 |
| chr8_136478593_136478847 | 0.13 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
7770 |
0.25 |
| chr19_57888956_57889107 | 0.13 |
ZNF547 |
zinc finger protein 547 |
6034 |
0.08 |
| chr1_215179577_215179737 | 0.13 |
KCNK2 |
potassium channel, subfamily K, member 2 |
459 |
0.91 |
| chr7_143581670_143581927 | 0.13 |
FAM115A |
family with sequence similarity 115, member A |
665 |
0.7 |
| chr3_167811230_167811432 | 0.13 |
GOLIM4 |
golgi integral membrane protein 4 |
1801 |
0.51 |
| chr2_170428489_170428760 | 0.13 |
FASTKD1 |
FAST kinase domains 1 |
875 |
0.59 |
| chr12_115250983_115251152 | 0.13 |
ENSG00000252459 |
. |
75558 |
0.12 |
| chr12_95084083_95084234 | 0.13 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
39820 |
0.19 |
| chr12_94132814_94133007 | 0.13 |
RP11-887P2.5 |
|
1311 |
0.51 |
| chr13_110496874_110497025 | 0.13 |
IRS2 |
insulin receptor substrate 2 |
58034 |
0.14 |
| chrX_109663875_109664026 | 0.13 |
RGAG1 |
retrotransposon gag domain containing 1 |
1665 |
0.42 |
| chr12_64239539_64239880 | 0.13 |
SRGAP1 |
SLIT-ROBO Rho GTPase activating protein 1 |
1168 |
0.42 |
| chr2_145276293_145276624 | 0.13 |
ZEB2-AS1 |
ZEB2 antisense RNA 1 |
434 |
0.52 |
| chr7_72570751_72570902 | 0.12 |
AC006995.8 |
|
45341 |
0.1 |
| chr8_4093405_4093556 | 0.12 |
ENSG00000264588 |
. |
156864 |
0.04 |
| chr4_186388513_186388728 | 0.12 |
RP11-279O9.4 |
Uncharacterized protein |
4050 |
0.16 |
| chr7_95239102_95239253 | 0.12 |
AC002451.3 |
|
69 |
0.98 |
| chr4_97329089_97329304 | 0.12 |
ENSG00000201640 |
. |
59007 |
0.17 |
| chr9_91608315_91608498 | 0.12 |
S1PR3 |
sphingosine-1-phosphate receptor 3 |
2044 |
0.31 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.0 | 0.0 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0035929 | steroid hormone secretion(GO:0035929) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.3 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.4 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0090266 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.0 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0051138 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.0 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.2 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.3 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0032356 | single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |