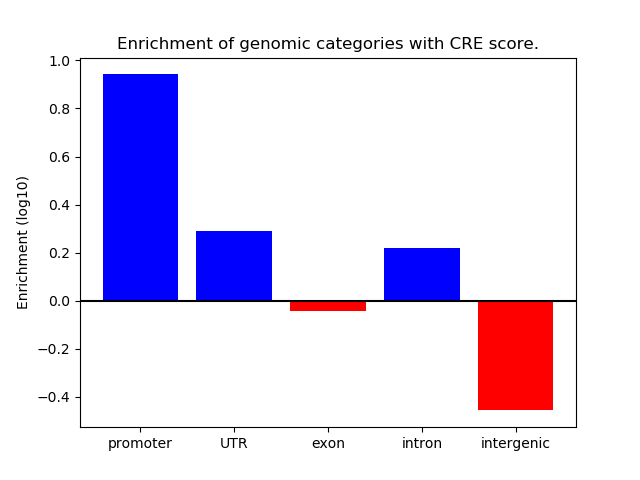
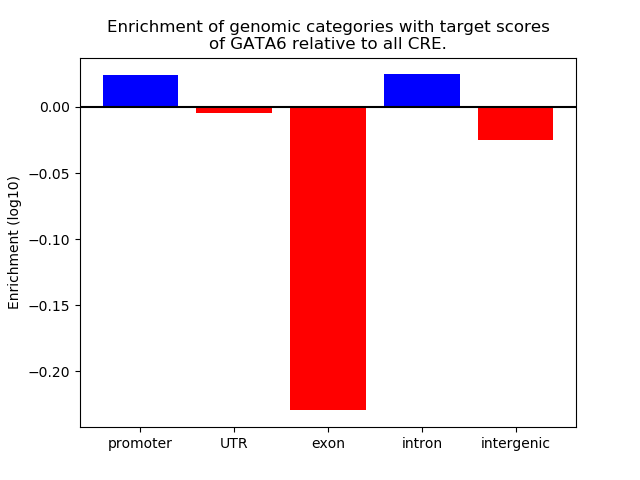
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
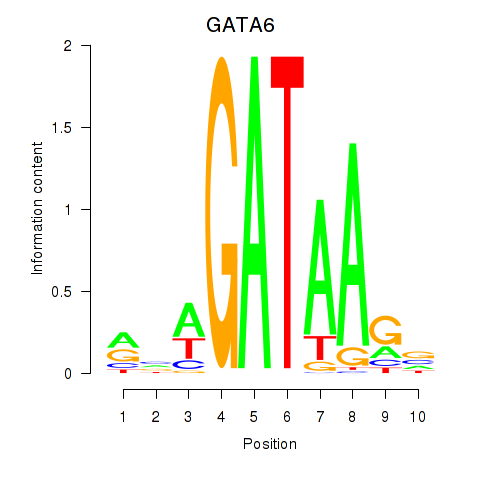
Results for GATA6
Z-value: 0.20
Transcription factors associated with GATA6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA6
|
ENSG00000141448.4 | GATA binding protein 6 |
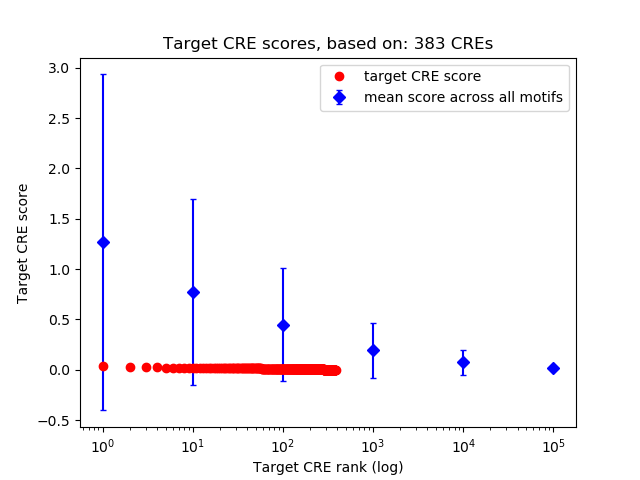
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_19751318_19751469 | GATA6 | 499 | 0.731776 | 0.84 | 4.6e-03 | Click! |
| chr18_19750904_19751196 | GATA6 | 156 | 0.938007 | 0.79 | 1.1e-02 | Click! |
| chr18_19747454_19747605 | GATA6 | 1875 | 0.266572 | 0.78 | 1.4e-02 | Click! |
| chr18_19749604_19749975 | GATA6 | 385 | 0.811827 | 0.73 | 2.6e-02 | Click! |
| chr18_19751508_19751659 | GATA6 | 689 | 0.608823 | 0.68 | 4.4e-02 | Click! |
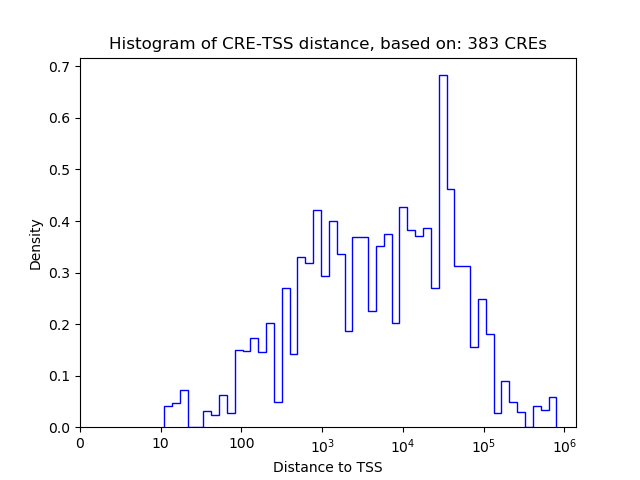
Activity of the GATA6 motif across conditions
Conditions sorted by the z-value of the GATA6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
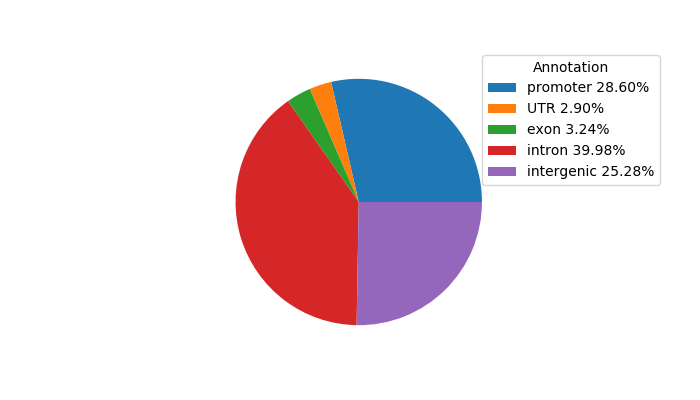
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_54074428_54074776 | 0.03 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
546 |
0.57 |
| chr14_75748202_75748353 | 0.03 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
1381 |
0.38 |
| chr9_21550613_21551072 | 0.02 |
MIR31HG |
MIR31 host gene (non-protein coding) |
8826 |
0.18 |
| chr7_116312981_116313724 | 0.02 |
MET |
met proto-oncogene |
893 |
0.68 |
| chr20_11871067_11871503 | 0.02 |
BTBD3 |
BTB (POZ) domain containing 3 |
86 |
0.98 |
| chr10_70817732_70817883 | 0.02 |
SRGN |
serglycin |
30067 |
0.15 |
| chr10_119124215_119124366 | 0.02 |
ENSG00000222197 |
. |
6423 |
0.22 |
| chr15_80747723_80747953 | 0.02 |
RP11-210M15.1 |
|
10345 |
0.19 |
| chr1_163039657_163039882 | 0.02 |
RGS4 |
regulator of G-protein signaling 4 |
618 |
0.82 |
| chr18_28783722_28783891 | 0.02 |
DSC1 |
desmocollin 1 |
40987 |
0.14 |
| chr12_123184919_123185107 | 0.02 |
HCAR2 |
hydroxycarboxylic acid receptor 2 |
2877 |
0.21 |
| chr22_46480171_46480322 | 0.02 |
FLJ27365 |
hsa-mir-4763 |
1637 |
0.21 |
| chr4_122706400_122706716 | 0.02 |
EXOSC9 |
exosome component 9 |
15914 |
0.15 |
| chr5_178712201_178712352 | 0.02 |
ADAMTS2 |
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
60155 |
0.14 |
| chr19_7460961_7461112 | 0.02 |
ARHGEF18 |
Rho/Rac guanine nucleotide exchange factor (GEF) 18 |
998 |
0.35 |
| chr2_122353801_122354268 | 0.02 |
ENSG00000222467 |
. |
6706 |
0.21 |
| chr3_187009488_187009765 | 0.02 |
MASP1 |
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
20 |
0.98 |
| chr4_122614681_122615038 | 0.02 |
ANXA5 |
annexin A5 |
3258 |
0.3 |
| chr8_33422706_33423010 | 0.02 |
RNF122 |
ring finger protein 122 |
1785 |
0.28 |
| chr12_88972313_88972545 | 0.02 |
KITLG |
KIT ligand |
1809 |
0.39 |
| chr3_98609388_98609782 | 0.02 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
10430 |
0.18 |
| chr21_38711458_38711729 | 0.02 |
AP001437.1 |
|
26641 |
0.16 |
| chr10_94180631_94180859 | 0.02 |
ENSG00000207895 |
. |
20316 |
0.21 |
| chr18_59660374_59660538 | 0.02 |
RNF152 |
ring finger protein 152 |
98992 |
0.08 |
| chr4_129243818_129244169 | 0.02 |
PGRMC2 |
progesterone receptor membrane component 2 |
34009 |
0.24 |
| chr16_8958188_8958339 | 0.02 |
CARHSP1 |
calcium regulated heat stable protein 1, 24kDa |
2629 |
0.18 |
| chr12_123198449_123198600 | 0.02 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
2915 |
0.19 |
| chr19_10764777_10764928 | 0.02 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
85 |
0.95 |
| chr16_84328151_84329018 | 0.02 |
WFDC1 |
WAP four-disulfide core domain 1 |
132 |
0.94 |
| chr7_130594902_130595108 | 0.02 |
ENSG00000226380 |
. |
32707 |
0.2 |
| chr7_87935974_87936151 | 0.02 |
STEAP4 |
STEAP family member 4 |
133 |
0.97 |
| chr5_17218970_17219230 | 0.02 |
BASP1 |
brain abundant, membrane attached signal protein 1 |
1431 |
0.35 |
| chr5_180677774_180677925 | 0.02 |
GNB2L1 |
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
2753 |
0.1 |
| chr6_108886554_108887901 | 0.02 |
FOXO3 |
forkhead box O3 |
5158 |
0.32 |
| chr2_8816989_8817431 | 0.02 |
AC011747.7 |
|
1254 |
0.45 |
| chr16_53906990_53907141 | 0.01 |
FTO |
fat mass and obesity associated |
13764 |
0.26 |
| chr1_181104582_181104733 | 0.01 |
IER5 |
immediate early response 5 |
47019 |
0.16 |
| chr12_8851550_8851701 | 0.01 |
RIMKLB |
ribosomal modification protein rimK-like family member B |
817 |
0.59 |
| chr7_92324656_92325109 | 0.01 |
ENSG00000206763 |
. |
6246 |
0.27 |
| chr4_126253702_126253855 | 0.01 |
FAT4 |
FAT atypical cadherin 4 |
16224 |
0.25 |
| chr2_197852750_197853145 | 0.01 |
ANKRD44 |
ankyrin repeat domain 44 |
12279 |
0.25 |
| chr1_234041439_234041955 | 0.01 |
SLC35F3 |
solute carrier family 35, member F3 |
1018 |
0.65 |
| chr17_58821066_58821723 | 0.01 |
ENSG00000252534 |
. |
5013 |
0.22 |
| chr8_25073004_25073155 | 0.01 |
DOCK5 |
dedicator of cytokinesis 5 |
30699 |
0.2 |
| chr16_10174074_10174418 | 0.01 |
RP11-895K13.2 |
|
46705 |
0.18 |
| chr7_134463594_134463964 | 0.01 |
CALD1 |
caldesmon 1 |
385 |
0.92 |
| chr12_46554202_46554353 | 0.01 |
SLC38A1 |
solute carrier family 38, member 1 |
107207 |
0.07 |
| chr1_79306379_79306727 | 0.01 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
77017 |
0.11 |
| chr10_14083265_14083416 | 0.01 |
FRMD4A |
FERM domain containing 4A |
32808 |
0.17 |
| chr2_24306708_24307223 | 0.01 |
TP53I3 |
tumor protein p53 inducible protein 3 |
216 |
0.89 |
| chr22_30176981_30177234 | 0.01 |
UQCR10 |
ubiquinol-cytochrome c reductase, complex III subunit X |
13744 |
0.12 |
| chr2_9350074_9350237 | 0.01 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
3261 |
0.37 |
| chr2_227767802_227767953 | 0.01 |
RHBDD1 |
rhomboid domain containing 1 |
3551 |
0.31 |
| chr9_137248904_137249055 | 0.01 |
ENSG00000263897 |
. |
22278 |
0.21 |
| chr7_114056620_114056771 | 0.01 |
FOXP2 |
forkhead box P2 |
1317 |
0.5 |
| chrX_46699952_46700103 | 0.01 |
RP2 |
retinitis pigmentosa 2 (X-linked recessive) |
3652 |
0.28 |
| chr6_44420303_44420454 | 0.01 |
ENSG00000266619 |
. |
17000 |
0.18 |
| chr7_134330596_134330773 | 0.01 |
BPGM |
2,3-bisphosphoglycerate mutase |
876 |
0.7 |
| chr3_114477625_114477831 | 0.01 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
59 |
0.98 |
| chr8_17600844_17600995 | 0.01 |
ENSG00000212280 |
. |
4708 |
0.19 |
| chr13_109624872_109625023 | 0.01 |
MYO16 |
myosin XVI |
86430 |
0.1 |
| chr8_70404668_70405416 | 0.01 |
SULF1 |
sulfatase 1 |
14 |
0.99 |
| chr5_138774627_138774795 | 0.01 |
DNAJC18 |
DnaJ (Hsp40) homolog, subfamily C, member 18 |
504 |
0.67 |
| chr17_57789761_57790061 | 0.01 |
VMP1 |
vacuole membrane protein 1 |
4828 |
0.18 |
| chr4_78124065_78124216 | 0.01 |
CCNG2 |
cyclin G2 |
44561 |
0.16 |
| chrX_151140603_151140985 | 0.01 |
GABRE |
gamma-aminobutyric acid (GABA) A receptor, epsilon |
2349 |
0.22 |
| chr2_217232127_217232304 | 0.01 |
MARCH4 |
membrane-associated ring finger (C3HC4) 4, E3 ubiquitin protein ligase |
4535 |
0.19 |
| chr8_93107061_93107485 | 0.01 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
170 |
0.98 |
| chr8_22516019_22516377 | 0.01 |
BIN3 |
bridging integrator 3 |
10428 |
0.11 |
| chr2_37870062_37870489 | 0.01 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
4526 |
0.29 |
| chr3_64174748_64175187 | 0.01 |
PRICKLE2-AS3 |
PRICKLE2 antisense RNA 3 |
1747 |
0.38 |
| chr11_118958508_118958659 | 0.01 |
HMBS |
hydroxymethylbilane synthase |
110 |
0.9 |
| chr5_59082098_59082249 | 0.01 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
17702 |
0.28 |
| chr3_128712183_128712559 | 0.01 |
KIAA1257 |
KIAA1257 |
548 |
0.71 |
| chr16_22307896_22308092 | 0.01 |
POLR3E |
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
736 |
0.63 |
| chr13_80065629_80065780 | 0.01 |
NDFIP2 |
Nedd4 family interacting protein 2 |
10116 |
0.21 |
| chr10_107734876_107735027 | 0.01 |
ENSG00000207068 |
. |
440534 |
0.01 |
| chr6_12324871_12325022 | 0.01 |
EDN1 |
endothelin 1 |
34350 |
0.22 |
| chr1_161593652_161593803 | 0.01 |
FCGR3B |
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
7095 |
0.12 |
| chr10_29607372_29607594 | 0.01 |
LYZL1 |
lysozyme-like 1 |
29493 |
0.23 |
| chr5_86409630_86409781 | 0.01 |
ENSG00000265919 |
. |
1066 |
0.58 |
| chr4_103605681_103605832 | 0.01 |
MANBA |
mannosidase, beta A, lysosomal |
76334 |
0.09 |
| chr4_168155836_168156001 | 0.01 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
10 |
0.99 |
| chr12_53896533_53896684 | 0.01 |
TARBP2 |
TAR (HIV-1) RNA binding protein 2 |
773 |
0.43 |
| chr10_121497247_121497413 | 0.01 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
11721 |
0.21 |
| chr6_106577027_106577713 | 0.01 |
RP1-134E15.3 |
|
29355 |
0.19 |
| chr7_87503839_87503990 | 0.01 |
DBF4 |
DBF4 homolog (S. cerevisiae) |
1630 |
0.34 |
| chr15_96936172_96936352 | 0.01 |
AC087477.1 |
Uncharacterized protein |
31775 |
0.16 |
| chr11_81727672_81727823 | 0.01 |
ENSG00000264110 |
. |
125869 |
0.06 |
| chr2_219152582_219152771 | 0.01 |
TMBIM1 |
transmembrane BAX inhibitor motif containing 1 |
650 |
0.54 |
| chr8_47819479_47819630 | 0.01 |
ENSG00000252594 |
. |
76882 |
0.12 |
| chr2_169952320_169952546 | 0.01 |
AC007556.3 |
|
4820 |
0.24 |
| chr3_162013497_162013648 | 0.01 |
OTOL1 |
otolin 1 |
798976 |
0.0 |
| chr7_6433170_6433329 | 0.01 |
RAC1 |
ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
19079 |
0.16 |
| chr5_59190026_59190177 | 0.01 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
476 |
0.89 |
| chr8_27629358_27630074 | 0.01 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
250 |
0.64 |
| chr9_132098034_132098185 | 0.01 |
ENSG00000242281 |
. |
34631 |
0.17 |
| chr6_153451642_153452011 | 0.01 |
RGS17 |
regulator of G-protein signaling 17 |
558 |
0.79 |
| chr4_13628130_13628980 | 0.01 |
BOD1L1 |
biorientation of chromosomes in cell division 1-like 1 |
792 |
0.46 |
| chr7_32243948_32244107 | 0.01 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
94914 |
0.09 |
| chr17_77001615_77001766 | 0.01 |
CANT1 |
calcium activated nucleotidase 1 |
4122 |
0.17 |
| chr10_31873172_31873323 | 0.01 |
ENSG00000222412 |
. |
171817 |
0.03 |
| chr10_24528419_24528570 | 0.01 |
KIAA1217 |
KIAA1217 |
370 |
0.91 |
| chr3_28337245_28337396 | 0.01 |
AZI2 |
5-azacytidine induced 2 |
36578 |
0.17 |
| chr15_63220786_63221067 | 0.01 |
RP11-1069G10.1 |
|
44693 |
0.15 |
| chr7_50564803_50564954 | 0.01 |
AC018705.5 |
|
34579 |
0.15 |
| chr14_54954818_54955652 | 0.01 |
GMFB |
glia maturation factor, beta |
141 |
0.96 |
| chr6_32120885_32121204 | 0.01 |
PPT2 |
palmitoyl-protein thioesterase 2 |
174 |
0.48 |
| chr6_71997364_71997515 | 0.01 |
OGFRL1 |
opioid growth factor receptor-like 1 |
1067 |
0.53 |
| chr16_67196775_67197002 | 0.01 |
HSF4 |
heat shock transcription factor 4 |
400 |
0.59 |
| chr1_68640038_68640228 | 0.01 |
ENSG00000221203 |
. |
9160 |
0.22 |
| chr5_134692650_134692921 | 0.01 |
C5orf66 |
chromosome 5 open reading frame 66 |
19156 |
0.17 |
| chr2_237600423_237600574 | 0.01 |
ACKR3 |
atypical chemokine receptor 3 |
122214 |
0.06 |
| chr9_89738562_89738713 | 0.01 |
C9orf170 |
chromosome 9 open reading frame 170 |
24922 |
0.26 |
| chr12_11948851_11949002 | 0.01 |
ETV6 |
ets variant 6 |
43491 |
0.19 |
| chr15_35600951_35601102 | 0.01 |
ENSG00000265102 |
. |
63539 |
0.15 |
| chr1_65453023_65453174 | 0.01 |
JAK1 |
Janus kinase 1 |
20911 |
0.19 |
| chr2_3527046_3527197 | 0.01 |
ADI1 |
acireductone dioxygenase 1 |
3614 |
0.17 |
| chr1_161136284_161136792 | 0.01 |
PPOX |
protoporphyrinogen oxidase |
237 |
0.79 |
| chr14_55915419_55915570 | 0.01 |
TBPL2 |
TATA box binding protein like 2 |
7950 |
0.23 |
| chr22_33141802_33142090 | 0.01 |
LL22NC01-116C6.1 |
|
37217 |
0.18 |
| chr4_159593239_159593390 | 0.01 |
ETFDH |
electron-transferring-flavoprotein dehydrogenase |
37 |
0.59 |
| chr6_143193241_143193570 | 0.01 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
35221 |
0.21 |
| chr8_103665091_103665311 | 0.01 |
KLF10 |
Kruppel-like factor 10 |
1402 |
0.49 |
| chr4_87898598_87898749 | 0.01 |
AFF1 |
AF4/FMR2 family, member 1 |
29467 |
0.2 |
| chr2_143631756_143631956 | 0.01 |
KYNU |
kynureninase |
3211 |
0.39 |
| chr2_111229667_111229818 | 0.01 |
LIMS3 |
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein |
651 |
0.43 |
| chr2_30670110_30671067 | 0.01 |
LCLAT1 |
lysocardiolipin acyltransferase 1 |
319 |
0.92 |
| chr8_96236254_96236405 | 0.01 |
C8orf37 |
chromosome 8 open reading frame 37 |
45100 |
0.16 |
| chr15_67992319_67992470 | 0.01 |
MAP2K5 |
mitogen-activated protein kinase kinase 5 |
59503 |
0.14 |
| chr6_26327913_26328565 | 0.01 |
ENSG00000252399 |
. |
25025 |
0.08 |
| chr4_82487360_82487596 | 0.01 |
RASGEF1B |
RasGEF domain family, member 1B |
94409 |
0.09 |
| chr15_35331378_35331529 | 0.01 |
ZNF770 |
zinc finger protein 770 |
50965 |
0.14 |
| chr6_37790580_37790731 | 0.01 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
2937 |
0.24 |
| chr22_31618868_31619019 | 0.01 |
ENSG00000199695 |
. |
104 |
0.94 |
| chr2_25599883_25600214 | 0.01 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
34589 |
0.16 |
| chr4_70778543_70778694 | 0.01 |
CSN1S1 |
casein alpha s1 |
18181 |
0.18 |
| chr4_10182385_10182585 | 0.01 |
WDR1 |
WD repeat domain 1 |
63912 |
0.11 |
| chr3_14211699_14211850 | 0.01 |
LSM3 |
LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
8084 |
0.16 |
| chr18_3306098_3306249 | 0.01 |
MYL12B |
myosin, light chain 12B, regulatory |
43219 |
0.12 |
| chr3_104552670_104552821 | 0.01 |
ALCAM |
activated leukocyte cell adhesion molecule |
533008 |
0.0 |
| chr6_138147763_138147914 | 0.01 |
RP11-356I2.4 |
|
807 |
0.68 |
| chr17_15159258_15159409 | 0.01 |
ENSG00000265110 |
. |
4320 |
0.16 |
| chr10_35595852_35596003 | 0.01 |
RP11-324I22.4 |
|
13999 |
0.18 |
| chr19_34216636_34216787 | 0.01 |
CHST8 |
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 8 |
41277 |
0.18 |
| chr17_38463018_38463169 | 0.01 |
RARA |
retinoic acid receptor, alpha |
2351 |
0.2 |
| chr12_27093586_27093737 | 0.01 |
FGFR1OP2 |
FGFR1 oncogene partner 2 |
2228 |
0.23 |
| chr14_70227453_70227703 | 0.01 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
6232 |
0.23 |
| chr16_84336242_84336393 | 0.01 |
RP11-558A11.2 |
|
5726 |
0.16 |
| chr6_111794162_111794313 | 0.01 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
10177 |
0.2 |
| chr10_70096270_70096993 | 0.01 |
PBLD |
phenazine biosynthesis-like protein domain containing |
3825 |
0.2 |
| chr9_89598436_89598587 | 0.01 |
GAS1 |
growth arrest-specific 1 |
36407 |
0.23 |
| chr11_5617858_5618363 | 0.01 |
TRIM6 |
tripartite motif containing 6 |
74 |
0.6 |
| chr6_132452290_132452699 | 0.01 |
ENSG00000265669 |
. |
16091 |
0.27 |
| chr11_34607506_34607749 | 0.01 |
EHF |
ets homologous factor |
35031 |
0.19 |
| chr1_177151039_177151190 | 0.01 |
ENSG00000222749 |
. |
4474 |
0.23 |
| chr18_46460407_46461027 | 0.01 |
SMAD7 |
SMAD family member 7 |
14158 |
0.24 |
| chr18_42746929_42747122 | 0.01 |
RP11-846C15.2 |
|
14054 |
0.26 |
| chr18_74826722_74826987 | 0.01 |
MBP |
myelin basic protein |
9637 |
0.3 |
| chr1_34368317_34368468 | 0.01 |
RP5-1007G16.1 |
|
27325 |
0.2 |
| chr11_109293842_109294216 | 0.01 |
C11orf87 |
chromosome 11 open reading frame 87 |
1183 |
0.65 |
| chr2_242479985_242480602 | 0.01 |
BOK |
BCL2-related ovarian killer |
17843 |
0.12 |
| chr10_77190038_77190238 | 0.01 |
RP11-399K21.10 |
|
1208 |
0.5 |
| chr6_56598112_56598263 | 0.01 |
DST |
dystonin |
52490 |
0.16 |
| chrX_123161918_123162069 | 0.01 |
STAG2 |
stromal antigen 2 |
64973 |
0.12 |
| chr15_96590422_96590657 | 0.01 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
175745 |
0.03 |
| chr2_110656312_110656910 | 0.01 |
LIMS3 |
LIM and senescent cell antigen-like domains 3 |
343 |
0.93 |
| chr12_122613090_122613241 | 0.01 |
MLXIP |
MLX interacting protein |
738 |
0.67 |
| chr6_36187187_36187338 | 0.01 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
15071 |
0.15 |
| chr2_28867267_28867418 | 0.01 |
PLB1 |
phospholipase B1 |
13889 |
0.18 |
| chr16_70779928_70780079 | 0.01 |
RP11-394B2.6 |
|
957 |
0.42 |
| chr12_129349404_129349555 | 0.01 |
GLT1D1 |
glycosyltransferase 1 domain containing 1 |
798 |
0.73 |
| chr1_201952184_201952974 | 0.01 |
RNPEP |
arginyl aminopeptidase (aminopeptidase B) |
121 |
0.93 |
| chr11_100719851_100720002 | 0.01 |
ENSG00000200047 |
. |
9565 |
0.23 |
| chr4_155728508_155728667 | 0.01 |
ENSG00000252604 |
. |
17144 |
0.15 |
| chr4_97494543_97494694 | 0.01 |
ENSG00000201640 |
. |
224429 |
0.02 |
| chr8_17220397_17220671 | 0.01 |
MTMR7 |
myotubularin related protein 7 |
50302 |
0.15 |
| chr1_181122215_181122496 | 0.01 |
IER5 |
immediate early response 5 |
64717 |
0.12 |
| chr2_85199843_85200208 | 0.01 |
KCMF1 |
potassium channel modulatory factor 1 |
701 |
0.74 |
| chr2_153349112_153349263 | 0.01 |
FMNL2 |
formin-like 2 |
126905 |
0.06 |
| chr12_6959922_6960405 | 0.01 |
CDCA3 |
cell division cycle associated 3 |
250 |
0.75 |
| chr13_96332061_96332487 | 0.01 |
DNAJC3 |
DnaJ (Hsp40) homolog, subfamily C, member 3 |
2862 |
0.29 |
| chr1_65886473_65887602 | 0.01 |
LEPR |
leptin receptor |
634 |
0.48 |
| chr3_72092545_72092696 | 0.01 |
ENSG00000239250 |
. |
216292 |
0.02 |
| chr15_89348318_89348469 | 0.01 |
ACAN |
aggrecan |
1403 |
0.51 |
| chr5_14026093_14026244 | 0.01 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
81516 |
0.11 |
| chrX_39921638_39921789 | 0.01 |
BCOR |
BCL6 corepressor |
477 |
0.9 |
| chr12_76100609_76100760 | 0.01 |
ENSG00000251893 |
. |
17600 |
0.22 |
| chr13_111365021_111365232 | 0.01 |
ING1 |
inhibitor of growth family, member 1 |
43 |
0.98 |
| chr13_99193927_99194078 | 0.01 |
STK24 |
serine/threonine kinase 24 |
19750 |
0.2 |
| chr8_28565361_28565512 | 0.01 |
EXTL3 |
exostosin-like glycosyltransferase 3 |
2439 |
0.28 |
| chr1_52113963_52114114 | 0.01 |
OSBPL9 |
oxysterol binding protein-like 9 |
21185 |
0.19 |
| chr7_23513086_23513237 | 0.01 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
3075 |
0.22 |
| chr3_23960351_23960502 | 0.01 |
RPL15 |
ribosomal protein L15 |
1230 |
0.41 |
| chrX_151999580_152000195 | 0.01 |
NSDHL |
NAD(P) dependent steroid dehydrogenase-like |
346 |
0.63 |
| chr1_27125657_27125808 | 0.01 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
10313 |
0.11 |
| chr1_23610032_23610183 | 0.01 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
60648 |
0.08 |
| chr15_86098904_86099149 | 0.01 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
349 |
0.88 |
| chr16_57798470_57798621 | 0.01 |
KIFC3 |
kinesin family member C3 |
244 |
0.9 |
| chr11_78356519_78356693 | 0.01 |
TENM4 |
teneurin transmembrane protein 4 |
56443 |
0.12 |
Network of associatons between targets according to the STRING database.