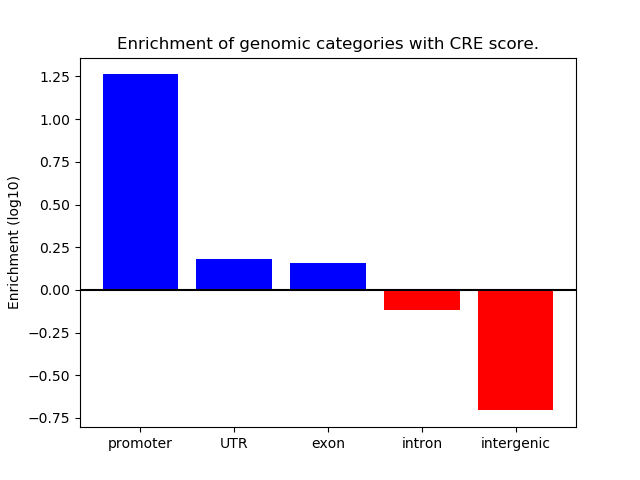
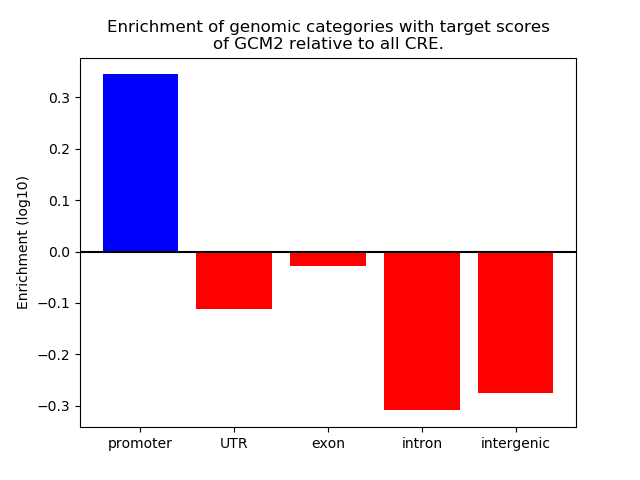
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
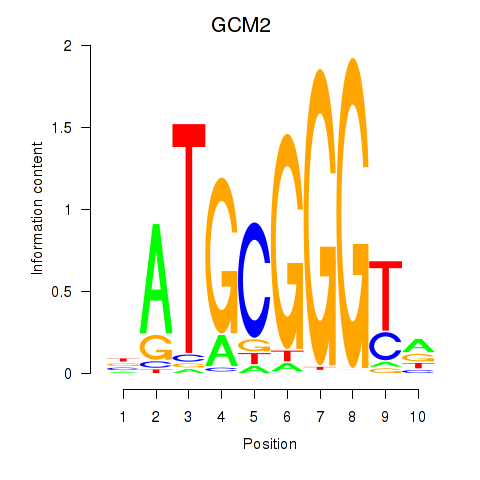
Results for GCM2
Z-value: 1.64
Transcription factors associated with GCM2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GCM2
|
ENSG00000124827.6 | glial cells missing transcription factor 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_10882334_10882485 | GCM2 | 235 | 0.655479 | 0.42 | 2.6e-01 | Click! |
| chr6_10882168_10882319 | GCM2 | 69 | 0.717054 | -0.42 | 2.6e-01 | Click! |
| chr6_10884165_10884460 | GCM2 | 2138 | 0.218969 | 0.10 | 7.9e-01 | Click! |
Activity of the GCM2 motif across conditions
Conditions sorted by the z-value of the GCM2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_38721330_38721617 | 0.74 |
CCR7 |
chemokine (C-C motif) receptor 7 |
238 |
0.92 |
| chr18_32556796_32556947 | 0.68 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
21 |
0.99 |
| chr19_18388821_18388972 | 0.59 |
JUND |
jun D proto-oncogene |
2843 |
0.13 |
| chr1_47799579_47799893 | 0.58 |
CMPK1 |
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
150 |
0.95 |
| chr10_11206834_11206985 | 0.55 |
CELF2 |
CUGBP, Elav-like family member 2 |
84 |
0.98 |
| chr8_144358025_144358176 | 0.50 |
RP13-582O9.5 |
|
4919 |
0.11 |
| chr14_23306583_23306834 | 0.49 |
MMP14 |
matrix metallopeptidase 14 (membrane-inserted) |
187 |
0.87 |
| chr5_179286017_179286230 | 0.46 |
CTC-241N9.1 |
Uncharacterized protein |
110 |
0.66 |
| chr7_116593207_116593413 | 0.46 |
ST7 |
suppression of tumorigenicity 7 |
18 |
0.9 |
| chr1_78245652_78246225 | 0.45 |
FAM73A |
family with sequence similarity 73, member A |
345 |
0.48 |
| chr17_5974794_5974945 | 0.45 |
WSCD1 |
WSC domain containing 1 |
441 |
0.9 |
| chr19_47127517_47127701 | 0.45 |
PTGIR |
prostaglandin I2 (prostacyclin) receptor (IP) |
685 |
0.49 |
| chr4_17578823_17578986 | 0.44 |
LAP3 |
leucine aminopeptidase 3 |
89 |
0.96 |
| chr17_37558067_37558428 | 0.44 |
CTB-131K11.1 |
|
201 |
0.65 |
| chr16_1032247_1032398 | 0.44 |
SOX8 |
SRY (sex determining region Y)-box 8 |
514 |
0.52 |
| chr8_116678670_116678821 | 0.43 |
TRPS1 |
trichorhinophalangeal syndrome I |
1463 |
0.59 |
| chr1_38512756_38512948 | 0.42 |
POU3F1 |
POU class 3 homeobox 1 |
402 |
0.82 |
| chr7_72300733_72300925 | 0.42 |
SBDSP1 |
Shwachman-Bodian-Diamond syndrome pseudogene 1 |
594 |
0.6 |
| chr7_5336962_5337158 | 0.42 |
SLC29A4 |
solute carrier family 29 (equilibrative nucleoside transporter), member 4 |
14400 |
0.16 |
| chr7_145813457_145813835 | 0.41 |
CNTNAP2 |
contactin associated protein-like 2 |
193 |
0.96 |
| chrX_134049034_134049244 | 0.41 |
MOSPD1 |
motile sperm domain containing 1 |
118 |
0.96 |
| chr5_159626613_159626831 | 0.41 |
CTB-127C13.1 |
|
620 |
0.46 |
| chr22_29195834_29196065 | 0.41 |
XBP1 |
X-box binding protein 1 |
172 |
0.86 |
| chrX_70287985_70288182 | 0.40 |
SNX12 |
sorting nexin 12 |
170 |
0.92 |
| chr19_23869476_23869665 | 0.40 |
ZNF675 |
zinc finger protein 675 |
387 |
0.9 |
| chr16_2015338_2015613 | 0.39 |
ENSG00000255198 |
. |
290 |
0.55 |
| chr10_104210626_104210853 | 0.39 |
ENSG00000269609 |
. |
351 |
0.57 |
| chr16_50583580_50583731 | 0.39 |
NKD1 |
naked cuticle homolog 1 (Drosophila) |
1414 |
0.38 |
| chrX_130937250_130937549 | 0.38 |
ENSG00000200587 |
. |
135585 |
0.05 |
| chr20_30948041_30948438 | 0.37 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
689 |
0.71 |
| chr3_46934468_46934619 | 0.36 |
AC109583.1 |
Uncharacterized protein |
4372 |
0.16 |
| chr6_26571959_26572122 | 0.36 |
ABT1 |
activator of basal transcription 1 |
25140 |
0.13 |
| chr17_40119579_40119751 | 0.36 |
CNP |
2',3'-cyclic nucleotide 3' phosphodiesterase |
136 |
0.94 |
| chr19_21688744_21688977 | 0.36 |
ZNF429 |
zinc finger protein 429 |
454 |
0.87 |
| chr20_62715149_62715509 | 0.35 |
C20orf201 |
chromosome 20 open reading frame 201 |
383 |
0.66 |
| chr6_34393564_34393808 | 0.35 |
RPS10-NUDT3 |
RPS10-NUDT3 readthrough |
139 |
0.61 |
| chr1_61548685_61548897 | 0.35 |
NFIA |
nuclear factor I/A |
365 |
0.52 |
| chr17_17643309_17643522 | 0.34 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
30720 |
0.12 |
| chr16_29623643_29623977 | 0.34 |
ENSG00000266758 |
. |
13224 |
0.14 |
| chr7_126988121_126988299 | 0.34 |
AC000123.4 |
|
1972 |
0.4 |
| chr6_18397050_18397268 | 0.33 |
ENSG00000238458 |
. |
5124 |
0.24 |
| chr15_93446819_93447199 | 0.33 |
ENSG00000264173 |
. |
620 |
0.48 |
| chr12_115122776_115122995 | 0.33 |
TBX3 |
T-box 3 |
916 |
0.64 |
| chr19_10531455_10531757 | 0.33 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
275 |
0.75 |
| chr10_28491840_28491991 | 0.33 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
35751 |
0.21 |
| chr15_89921416_89921567 | 0.33 |
ENSG00000207819 |
. |
10243 |
0.19 |
| chr17_34891354_34891543 | 0.32 |
PIGW |
phosphatidylinositol glycan anchor biosynthesis, class W |
45 |
0.91 |
| chr13_41886064_41886695 | 0.32 |
NAA16 |
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
963 |
0.57 |
| chr7_148396048_148396355 | 0.32 |
CUL1 |
cullin 1 |
197 |
0.75 |
| chr5_177027484_177027704 | 0.32 |
B4GALT7 |
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 |
363 |
0.8 |
| chr11_107328720_107328953 | 0.31 |
CWF19L2 |
CWF19-like 2, cell cycle control (S. pombe) |
264 |
0.95 |
| chr6_131456080_131456669 | 0.31 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
432 |
0.9 |
| chr5_73982840_73982991 | 0.31 |
HEXB |
hexosaminidase B (beta polypeptide) |
1946 |
0.34 |
| chr17_16256516_16256704 | 0.30 |
CENPV |
centromere protein V |
160 |
0.94 |
| chr1_153963262_153963499 | 0.30 |
RPS27 |
ribosomal protein S27 |
142 |
0.9 |
| chr6_26033320_26033579 | 0.30 |
HIST1H2AB |
histone cluster 1, H2ab |
347 |
0.65 |
| chr5_96271059_96271537 | 0.30 |
LNPEP |
leucyl/cystinyl aminopeptidase |
130 |
0.96 |
| chr2_26569875_26570036 | 0.30 |
GPR113 |
G protein-coupled receptor 113 |
270 |
0.84 |
| chr3_53916371_53916541 | 0.30 |
ENSG00000222888 |
. |
160 |
0.58 |
| chr9_131903521_131903756 | 0.30 |
PPP2R4 |
protein phosphatase 2A activator, regulatory subunit 4 |
287 |
0.85 |
| chr10_104170136_104170328 | 0.30 |
PSD |
pleckstrin and Sec7 domain containing |
8669 |
0.1 |
| chrX_1572414_1572620 | 0.30 |
ASMTL |
acetylserotonin O-methyltransferase-like |
138 |
0.96 |
| chr19_53104022_53104358 | 0.30 |
CTD-3099C6.11 |
|
2050 |
0.19 |
| chr4_57931934_57932158 | 0.29 |
ENSG00000238541 |
. |
40465 |
0.11 |
| chr7_143533030_143533370 | 0.29 |
RP11-61L23.2 |
|
104 |
0.97 |
| chr3_42846205_42846412 | 0.29 |
HIGD1A |
HIG1 hypoxia inducible domain family, member 1A |
285 |
0.84 |
| chr11_100558368_100558828 | 0.29 |
CTD-2383M3.1 |
|
88 |
0.65 |
| chrY_1522413_1522621 | 0.29 |
NA |
NA |
> 106 |
NA |
| chr13_22057996_22058198 | 0.29 |
ZDHHC20 |
zinc finger, DHHC-type containing 20 |
24588 |
0.18 |
| chr7_195524_195675 | 0.29 |
AC093627.12 |
|
1419 |
0.41 |
| chr7_143318509_143318843 | 0.29 |
FAM115C |
family with sequence similarity 115, member C |
131 |
0.96 |
| chr8_121744015_121744417 | 0.29 |
RP11-713M15.1 |
|
29277 |
0.22 |
| chr19_47634546_47634940 | 0.29 |
SAE1 |
SUMO1 activating enzyme subunit 1 |
591 |
0.71 |
| chr6_169990007_169990169 | 0.29 |
WDR27 |
WD repeat domain 27 |
70700 |
0.1 |
| chr18_68326306_68326457 | 0.29 |
GTSCR1 |
Gilles de la Tourette syndrome chromosome region, candidate 1 |
8401 |
0.32 |
| chr17_65241489_65241782 | 0.29 |
HELZ |
helicase with zinc finger |
332 |
0.69 |
| chr3_119014697_119015414 | 0.29 |
ARHGAP31 |
Rho GTPase activating protein 31 |
1835 |
0.31 |
| chr6_26032878_26033311 | 0.28 |
HIST1H2AB |
histone cluster 1, H2ab |
702 |
0.31 |
| chr22_41940614_41940785 | 0.28 |
POLR3H |
polymerase (RNA) III (DNA directed) polypeptide H (22.9kD) |
89 |
0.96 |
| chr2_101180136_101180337 | 0.28 |
PDCL3 |
phosducin-like 3 |
516 |
0.76 |
| chr3_183966275_183966684 | 0.28 |
ALG3 |
ALG3, alpha-1,3- mannosyltransferase |
244 |
0.79 |
| chr17_79935874_79936121 | 0.28 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
222 |
0.82 |
| chr19_15343730_15343881 | 0.28 |
EPHX3 |
epoxide hydrolase 3 |
53 |
0.97 |
| chr22_50243479_50243664 | 0.28 |
ZBED4 |
zinc finger, BED-type containing 4 |
3919 |
0.22 |
| chr11_35965131_35965474 | 0.28 |
LDLRAD3 |
low density lipoprotein receptor class A domain containing 3 |
229 |
0.93 |
| chr9_116163187_116163544 | 0.28 |
ALAD |
aminolevulinate dehydratase |
35 |
0.96 |
| chr6_26198818_26199250 | 0.28 |
HIST1H2AD |
histone cluster 1, H2ad |
437 |
0.34 |
| chr21_33244491_33244642 | 0.28 |
HUNK |
hormonally up-regulated Neu-associated kinase |
1062 |
0.65 |
| chr3_143566573_143567346 | 0.28 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
341 |
0.94 |
| chr12_32871598_32871749 | 0.28 |
ENSG00000201563 |
. |
13233 |
0.21 |
| chr2_174829537_174830027 | 0.28 |
SP3 |
Sp3 transcription factor |
501 |
0.89 |
| chr14_23299131_23299994 | 0.28 |
MRPL52 |
mitochondrial ribosomal protein L52 |
304 |
0.6 |
| chr17_7164973_7165404 | 0.27 |
CLDN7 |
claudin 7 |
474 |
0.52 |
| chr7_99102295_99102499 | 0.27 |
ZKSCAN5 |
zinc finger with KRAB and SCAN domains 5 |
99 |
0.94 |
| chrX_1514201_1514352 | 0.27 |
SLC25A6 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
2659 |
0.21 |
| chr11_65154078_65154899 | 0.27 |
FRMD8 |
FERM domain containing 8 |
89 |
0.94 |
| chr10_99093753_99093904 | 0.27 |
RP11-452K12.4 |
|
493 |
0.49 |
| chr2_191745621_191745919 | 0.27 |
GLS |
glutaminase |
210 |
0.9 |
| chr11_66035600_66035751 | 0.27 |
RP11-867G23.2 |
|
75 |
0.76 |
| chr16_29823564_29823949 | 0.27 |
PRRT2 |
proline-rich transmembrane protein 2 |
192 |
0.68 |
| chrY_1464199_1464350 | 0.27 |
NA |
NA |
> 106 |
NA |
| chr16_86604479_86604706 | 0.27 |
RP11-463O9.5 |
|
3225 |
0.19 |
| chrX_48693575_48693797 | 0.27 |
PCSK1N |
proprotein convertase subtilisin/kexin type 1 inhibitor |
349 |
0.78 |
| chr1_16375359_16375546 | 0.27 |
CLCNKB |
chloride channel, voltage-sensitive Kb |
168 |
0.92 |
| chr12_25956104_25956320 | 0.27 |
ENSG00000222950 |
. |
23709 |
0.25 |
| chr16_82661012_82661382 | 0.26 |
CDH13 |
cadherin 13 |
499 |
0.88 |
| chr1_54844833_54844984 | 0.26 |
SSBP3 |
single stranded DNA binding protein 3 |
26269 |
0.19 |
| chrX_153084272_153084494 | 0.26 |
PDZD4 |
PDZ domain containing 4 |
10968 |
0.09 |
| chr19_20278322_20278527 | 0.26 |
ZNF486 |
zinc finger protein 486 |
341 |
0.87 |
| chrX_53024151_53024564 | 0.26 |
FAM156A |
family with sequence similarity 156, member A |
194 |
0.95 |
| chr15_49103006_49103287 | 0.26 |
CEP152 |
centrosomal protein 152kDa |
38 |
0.76 |
| chr2_230873136_230873287 | 0.26 |
ENSG00000206725 |
. |
35180 |
0.13 |
| chr22_51039548_51039740 | 0.26 |
MAPK8IP2 |
mitogen-activated protein kinase 8 interacting protein 2 |
513 |
0.6 |
| chr1_116711224_116711506 | 0.26 |
MAB21L3 |
mab-21-like 3 (C. elegans) |
56989 |
0.14 |
| chr11_307271_307422 | 0.26 |
IFITM2 |
interferon induced transmembrane protein 2 |
285 |
0.75 |
| chr5_36878186_36878337 | 0.25 |
NIPBL |
Nipped-B homolog (Drosophila) |
1369 |
0.59 |
| chr5_160974690_160974841 | 0.25 |
GABRB2 |
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
365 |
0.93 |
| chr4_27050261_27050494 | 0.25 |
STIM2 |
stromal interaction molecule 2 |
40282 |
0.22 |
| chr2_233387084_233387235 | 0.25 |
PRSS56 |
protease, serine, 56 |
1986 |
0.18 |
| chr17_48619640_48620019 | 0.25 |
ENSG00000250286 |
. |
3714 |
0.11 |
| chrX_9320798_9321168 | 0.25 |
TBL1X |
transducin (beta)-like 1X-linked |
110352 |
0.07 |
| chr15_74219539_74219761 | 0.25 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
386 |
0.69 |
| chr7_100859878_100860119 | 0.25 |
PLOD3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
697 |
0.4 |
| chr11_66386268_66386833 | 0.25 |
RBM14 |
RNA binding motif protein 14 |
2364 |
0.12 |
| chr1_10532708_10532986 | 0.25 |
DFFA |
DNA fragmentation factor, 45kDa, alpha polypeptide |
264 |
0.85 |
| chr19_53466333_53466748 | 0.25 |
ZNF816 |
zinc finger protein 816 |
376 |
0.53 |
| chr20_3788549_3788700 | 0.25 |
CDC25B |
cell division cycle 25B |
11546 |
0.11 |
| chr14_90528183_90528389 | 0.25 |
KCNK13 |
potassium channel, subfamily K, member 13 |
177 |
0.97 |
| chr2_66664825_66665240 | 0.24 |
MEIS1 |
Meis homeobox 1 |
93 |
0.96 |
| chr6_25732828_25732979 | 0.24 |
HIST1H2BA |
histone cluster 1, H2ba |
5766 |
0.17 |
| chr5_176023977_176024277 | 0.24 |
GPRIN1 |
G protein regulated inducer of neurite outgrowth 1 |
13007 |
0.11 |
| chr20_43211231_43211439 | 0.24 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
145 |
0.95 |
| chr17_21030010_21030234 | 0.24 |
DHRS7B |
dehydrogenase/reductase (SDR family) member 7B |
155 |
0.96 |
| chr5_60921640_60922072 | 0.24 |
C5orf64 |
chromosome 5 open reading frame 64 |
11780 |
0.22 |
| chr8_136556013_136556164 | 0.24 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
1142 |
0.65 |
| chr5_39526290_39526441 | 0.24 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
63963 |
0.15 |
| chr9_4741404_4741742 | 0.24 |
AK3 |
adenylate kinase 3 |
255 |
0.92 |
| chr6_27805401_27805998 | 0.24 |
HIST1H2AK |
histone cluster 1, H2ak |
418 |
0.5 |
| chr5_158533512_158533971 | 0.24 |
EBF1 |
early B-cell factor 1 |
6972 |
0.26 |
| chr15_96952221_96952372 | 0.24 |
AC087477.1 |
Uncharacterized protein |
47809 |
0.13 |
| chr1_223536976_223537611 | 0.24 |
SUSD4 |
sushi domain containing 4 |
108 |
0.97 |
| chr17_42733796_42734027 | 0.24 |
C17orf104 |
chromosome 17 open reading frame 104 |
71 |
0.96 |
| chr2_220299665_220299816 | 0.24 |
SPEG |
SPEG complex locus |
172 |
0.9 |
| chr6_27101035_27101630 | 0.24 |
HIST1H2AG |
histone cluster 1, H2ag |
500 |
0.53 |
| chr7_66459685_66459882 | 0.24 |
SBDS |
Shwachman-Bodian-Diamond syndrome |
805 |
0.55 |
| chr21_44922813_44922964 | 0.24 |
SIK1 |
salt-inducible kinase 1 |
75880 |
0.1 |
| chr10_75646822_75647179 | 0.24 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
12657 |
0.12 |
| chr1_40562359_40562905 | 0.24 |
PPT1 |
palmitoyl-protein thioesterase 1 |
93 |
0.97 |
| chr11_46450627_46450944 | 0.24 |
AMBRA1 |
autophagy/beclin-1 regulator 1 |
11240 |
0.15 |
| chr22_36173418_36173584 | 0.23 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
578 |
0.82 |
| chr1_145472532_145473065 | 0.23 |
ANKRD34A |
ankyrin repeat domain 34A |
2290 |
0.15 |
| chr19_38397857_38398210 | 0.23 |
SIPA1L3 |
signal-induced proliferation-associated 1 like 3 |
165 |
0.87 |
| chr6_44356035_44356348 | 0.23 |
CDC5L |
cell division cycle 5-like |
929 |
0.54 |
| chr2_112895743_112895934 | 0.23 |
FBLN7 |
fibulin 7 |
124 |
0.97 |
| chr2_179344744_179345027 | 0.23 |
PLEKHA3 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
310 |
0.82 |
| chr15_60883034_60883482 | 0.23 |
RORA |
RAR-related orphan receptor A |
1482 |
0.47 |
| chr13_47159131_47159282 | 0.23 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
31811 |
0.22 |
| chr17_75372530_75372681 | 0.23 |
SEPT9 |
septin 9 |
440 |
0.62 |
| chr6_157935851_157936002 | 0.23 |
ENSG00000266617 |
. |
14238 |
0.27 |
| chr3_180320366_180320663 | 0.23 |
TTC14 |
tetratricopeptide repeat domain 14 |
164 |
0.95 |
| chr19_50393038_50393189 | 0.23 |
ENSG00000263462 |
. |
1681 |
0.14 |
| chr7_138665064_138665872 | 0.23 |
KIAA1549 |
KIAA1549 |
596 |
0.79 |
| chr19_21580209_21580417 | 0.23 |
ZNF493 |
zinc finger protein 493 |
345 |
0.9 |
| chr1_204485580_204485809 | 0.23 |
MDM4 |
Mdm4 p53 binding protein homolog (mouse) |
75 |
0.97 |
| chr21_33783968_33784413 | 0.23 |
EVA1C |
eva-1 homolog C (C. elegans) |
499 |
0.74 |
| chr17_56592126_56592438 | 0.23 |
MTMR4 |
myotubularin related protein 4 |
297 |
0.84 |
| chr7_149746137_149746288 | 0.22 |
ATP6V0E2-AS1 |
ATP6V0E2 antisense RNA 1 |
168513 |
0.03 |
| chr7_1177432_1177637 | 0.22 |
C7orf50 |
chromosome 7 open reading frame 50 |
342 |
0.83 |
| chr6_127587718_127587944 | 0.22 |
RNF146 |
ring finger protein 146 |
40 |
0.98 |
| chr12_118499916_118500430 | 0.22 |
WSB2 |
WD repeat and SOCS box containing 2 |
42 |
0.98 |
| chr10_3111439_3111730 | 0.22 |
PFKP |
phosphofructokinase, platelet |
765 |
0.76 |
| chr7_16686358_16686839 | 0.22 |
BZW2 |
basic leucine zipper and W2 domains 2 |
778 |
0.52 |
| chr2_196521999_196522280 | 0.22 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
34 |
0.98 |
| chr19_58361206_58361626 | 0.22 |
ZNF587 |
zinc finger protein 587 |
188 |
0.89 |
| chr19_44031024_44031309 | 0.22 |
ETHE1 |
ethylmalonic encephalopathy 1 |
187 |
0.87 |
| chr1_76189629_76189967 | 0.22 |
RP4-682C21.5 |
|
102 |
0.69 |
| chr17_48906674_48906825 | 0.22 |
WFIKKN2 |
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
5262 |
0.17 |
| chr7_76179763_76179914 | 0.22 |
AC004980.7 |
|
903 |
0.55 |
| chr15_69110393_69110544 | 0.22 |
ANP32A |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
2757 |
0.31 |
| chr2_232480061_232480212 | 0.22 |
C2orf57 |
chromosome 2 open reading frame 57 |
22561 |
0.15 |
| chr11_57103479_57104011 | 0.22 |
SSRP1 |
structure specific recognition protein 1 |
394 |
0.71 |
| chr19_21325116_21325348 | 0.22 |
ZNF431 |
zinc finger protein 431 |
352 |
0.89 |
| chr1_116184049_116184456 | 0.22 |
VANGL1 |
VANGL planar cell polarity protein 1 |
322 |
0.9 |
| chr9_35748225_35748926 | 0.22 |
RGP1 |
RGP1 retrograde golgi transport homolog (S. cerevisiae) |
628 |
0.34 |
| chr11_33060532_33060760 | 0.22 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
317 |
0.89 |
| chr4_189321790_189321972 | 0.22 |
RP11-366H4.3 |
|
257348 |
0.02 |
| chr5_140762259_140762453 | 0.21 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
111 |
0.88 |
| chr3_13590656_13590970 | 0.21 |
FBLN2 |
fibulin 2 |
132 |
0.97 |
| chr17_18528054_18528535 | 0.21 |
CCDC144B |
coiled-coil domain containing 144B (pseudogene) |
466 |
0.74 |
| chr6_31784437_31784624 | 0.21 |
HSPA1L |
heat shock 70kDa protein 1-like |
1093 |
0.17 |
| chr11_8710014_8710214 | 0.21 |
ENSG00000212607 |
. |
3128 |
0.13 |
| chr11_35440658_35441040 | 0.21 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
53 |
0.75 |
| chr10_126431828_126432166 | 0.21 |
FAM53B |
family with sequence similarity 53, member B |
458 |
0.8 |
| chr2_62423858_62424104 | 0.21 |
B3GNT2 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
733 |
0.65 |
| chr19_47365826_47365977 | 0.21 |
AP2S1 |
adaptor-related protein complex 2, sigma 1 subunit |
11652 |
0.17 |
| chr1_19277426_19277605 | 0.21 |
IFFO2 |
intermediate filament family orphan 2 |
4677 |
0.22 |
| chr4_39460235_39460455 | 0.21 |
RPL9 |
ribosomal protein L9 |
163 |
0.64 |
| chr1_1656054_1656334 | 0.21 |
CDK11A |
cyclin-dependent kinase 11A |
428 |
0.74 |
| chr17_65983_66134 | 0.21 |
DOC2B |
double C2-like domains, beta |
34631 |
0.16 |
| chr18_21243108_21243324 | 0.21 |
ANKRD29 |
ankyrin repeat domain 29 |
367 |
0.88 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.2 | 0.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.1 | 0.3 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
| 0.1 | 0.3 | GO:0072110 | glomerular mesangial cell proliferation(GO:0072110) regulation of glomerular mesangial cell proliferation(GO:0072124) |
| 0.1 | 0.4 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.1 | GO:0090192 | regulation of glomerulus development(GO:0090192) |
| 0.1 | 0.3 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.1 | 0.2 | GO:0072109 | glomerular mesangium development(GO:0072109) |
| 0.1 | 0.3 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.7 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.4 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.1 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 | 0.2 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.3 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 0.2 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.2 | GO:0061316 | negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) canonical Wnt signaling pathway involved in heart development(GO:0061316) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.1 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.4 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.1 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.1 | 0.2 | GO:0035751 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 0.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.2 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 0.2 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.2 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.1 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.1 | 0.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.2 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.1 | 0.3 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.1 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.1 | 0.3 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.1 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.2 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0000462 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) cleavage involved in rRNA processing(GO:0000469) endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.2 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0021561 | facial nerve development(GO:0021561) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.5 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.5 | GO:0006385 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.1 | GO:0035844 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.2 | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process(GO:0009129) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.2 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.2 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.2 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:1904376 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.2 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0052251 | induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0042424 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0014808 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.2 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.2 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.2 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.4 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:1903313 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.2 | GO:0032727 | positive regulation of interferon-alpha production(GO:0032727) |
| 0.0 | 0.0 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.0 | GO:0002674 | negative regulation of acute inflammatory response(GO:0002674) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.0 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.4 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0006228 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 | 0.3 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0003356 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.0 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0043558 | regulation of translational initiation in response to stress(GO:0043558) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) negative regulation of cell killing(GO:0031342) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway(GO:0033145) |
| 0.0 | 0.2 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.0 | 0.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.0 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.1 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.2 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 0.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.0 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.0 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.1 | GO:0042069 | regulation of catecholamine metabolic process(GO:0042069) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.2 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0097300 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.0 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 | 0.1 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) |
| 0.0 | 0.0 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.1 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) |
| 0.0 | 0.3 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.3 | GO:0050684 | regulation of mRNA processing(GO:0050684) regulation of mRNA metabolic process(GO:1903311) |
| 0.0 | 0.0 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 1.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.0 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0072577 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.1 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.0 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.0 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.0 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.2 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.0 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:0090494 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0042088 | T-helper 1 type immune response(GO:0042088) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.0 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.0 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.1 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.0 | GO:0010713 | negative regulation of collagen metabolic process(GO:0010713) negative regulation of collagen biosynthetic process(GO:0032966) negative regulation of multicellular organismal metabolic process(GO:0044252) |
| 0.0 | 0.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.0 | GO:0032648 | regulation of interferon-beta production(GO:0032648) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0032730 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:0070431 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.0 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.0 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 0.0 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:1902743 | regulation of lamellipodium assembly(GO:0010591) regulation of lamellipodium organization(GO:1902743) |
| 0.0 | 0.1 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.0 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.0 | GO:0048263 | determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.2 | GO:0006490 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.3 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.1 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0043230 | extracellular organelle(GO:0043230) extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0044217 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.3 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.6 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.0 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 1.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.2 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.2 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.1 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.3 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.2 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.0 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.0 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.2 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.0 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.0 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.0 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.0 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.3 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.0 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.0 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.0 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME SIGNALLING TO ERKS | Genes involved in Signalling to ERKs |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |