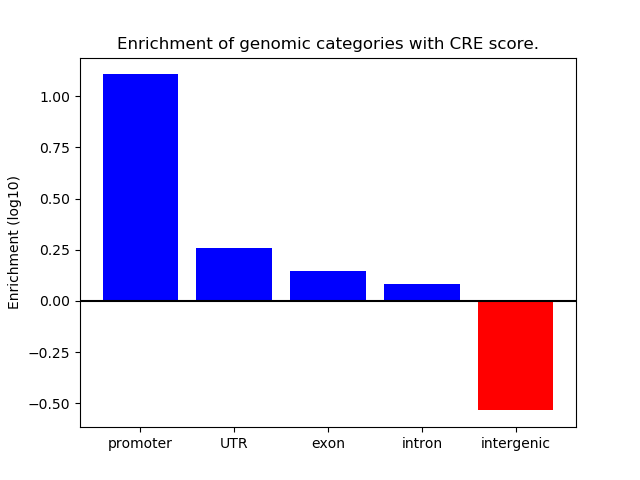
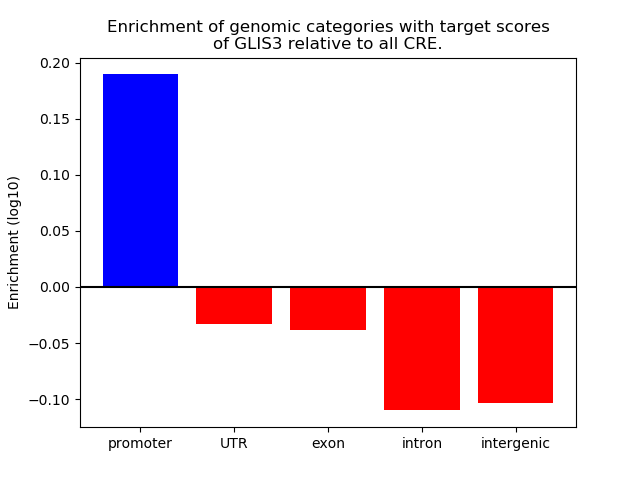
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
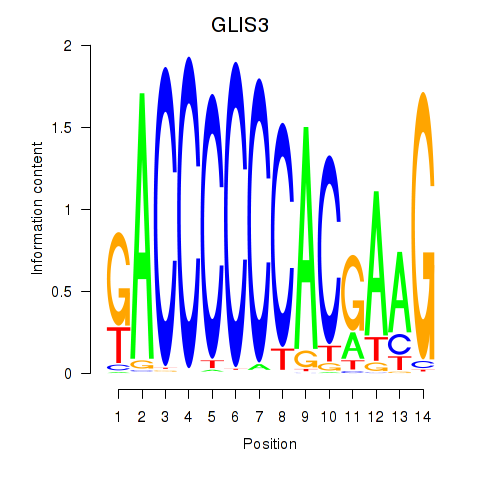
Results for GLIS3
Z-value: 0.74
Transcription factors associated with GLIS3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS3
|
ENSG00000107249.17 | GLIS family zinc finger 3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_4293611_4293830 | GLIS3 | 4776 | 0.234420 | 0.92 | 4.2e-04 | Click! |
| chr9_4301764_4301915 | GLIS3 | 1923 | 0.325141 | 0.91 | 5.8e-04 | Click! |
| chr9_4144974_4145179 | GLIS3 | 117 | 0.979043 | 0.89 | 1.5e-03 | Click! |
| chr9_4278077_4278228 | GLIS3 | 20344 | 0.199196 | 0.89 | 1.5e-03 | Click! |
| chr9_4225853_4226004 | GLIS3 | 72568 | 0.104218 | 0.86 | 2.9e-03 | Click! |
Activity of the GLIS3 motif across conditions
Conditions sorted by the z-value of the GLIS3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
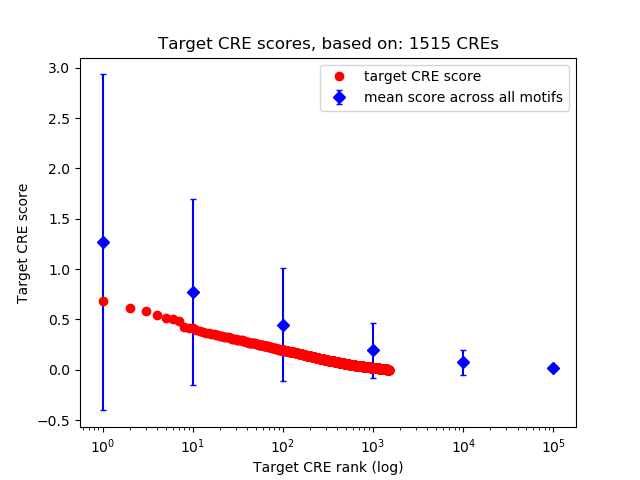
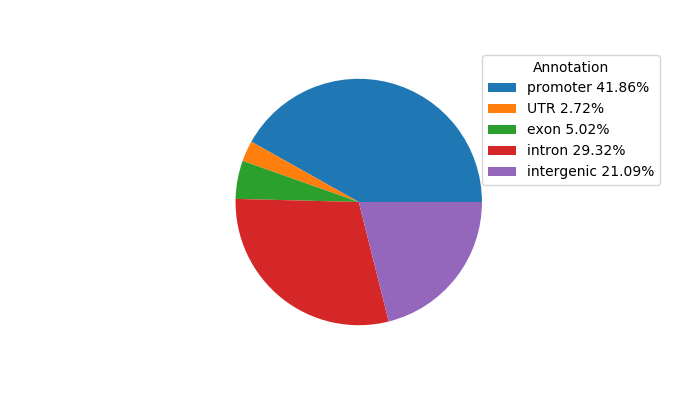
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_107178963_107179207 | 0.68 |
RP6-191P20.4 |
|
125 |
0.97 |
| chr17_59476045_59476898 | 0.61 |
RP11-332H18.4 |
|
260 |
0.74 |
| chr2_27717409_27717598 | 0.59 |
FNDC4 |
fibronectin type III domain containing 4 |
609 |
0.54 |
| chr2_190042287_190042516 | 0.54 |
COL5A2 |
collagen, type V, alpha 2 |
2204 |
0.37 |
| chr22_51136330_51136565 | 0.52 |
ENSG00000206841 |
. |
6656 |
0.12 |
| chr16_86601461_86601775 | 0.50 |
RP11-463O9.5 |
|
251 |
0.8 |
| chr2_121493751_121494304 | 0.49 |
GLI2 |
GLI family zinc finger 2 |
204 |
0.97 |
| chr3_48632919_48633070 | 0.42 |
COL7A1 |
collagen, type VII, alpha 1 |
294 |
0.8 |
| chr3_50193036_50193699 | 0.42 |
RP11-493K19.3 |
|
86 |
0.87 |
| chr7_71098294_71098514 | 0.41 |
ENSG00000199940 |
. |
52328 |
0.19 |
| chr5_140887996_140888228 | 0.40 |
DIAPH1 |
diaphanous-related formin 1 |
17602 |
0.08 |
| chr1_17867304_17867455 | 0.39 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
1049 |
0.62 |
| chr21_38071587_38071762 | 0.38 |
SIM2 |
single-minded family bHLH transcription factor 2 |
240 |
0.92 |
| chr2_236401822_236402641 | 0.37 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
502 |
0.83 |
| chr17_79397329_79397532 | 0.37 |
RP11-1055B8.7 |
BAH and coiled-coil domain-containing protein 1 |
7966 |
0.11 |
| chr14_29228686_29229081 | 0.36 |
RP11-966I7.1 |
|
5586 |
0.17 |
| chr20_6103412_6103655 | 0.36 |
FERMT1 |
fermitin family member 1 |
144 |
0.97 |
| chr8_494526_494820 | 0.34 |
TDRP |
testis development related protein |
164 |
0.97 |
| chr3_157260663_157261197 | 0.34 |
C3orf55 |
chromosome 3 open reading frame 55 |
105 |
0.98 |
| chr12_122467875_122468026 | 0.34 |
BCL7A |
B-cell CLL/lymphoma 7A |
8158 |
0.22 |
| chr13_110958781_110959436 | 0.34 |
COL4A1 |
collagen, type IV, alpha 1 |
370 |
0.6 |
| chr11_119597594_119597745 | 0.33 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
1594 |
0.37 |
| chr17_70120328_70120596 | 0.33 |
SOX9 |
SRY (sex determining region Y)-box 9 |
3301 |
0.38 |
| chr16_51184162_51184313 | 0.32 |
SALL1 |
spalt-like transcription factor 1 |
271 |
0.87 |
| chr22_44315340_44315603 | 0.32 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
4148 |
0.18 |
| chr22_43523181_43523332 | 0.31 |
MCAT |
malonyl CoA:ACP acyltransferase (mitochondrial) |
15714 |
0.13 |
| chr12_46886554_46886705 | 0.31 |
SLC38A2 |
solute carrier family 38, member 2 |
119979 |
0.06 |
| chr16_67215217_67215453 | 0.31 |
KIAA0895L |
KIAA0895-like |
1370 |
0.18 |
| chr19_16936008_16936179 | 0.31 |
SIN3B |
SIN3 transcription regulator family member B |
4118 |
0.16 |
| chr11_61739742_61739933 | 0.30 |
AP003733.1 |
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
4384 |
0.15 |
| chr8_11421540_11421831 | 0.30 |
RP11-148O21.2 |
|
4156 |
0.18 |
| chr1_186649638_186650434 | 0.30 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
477 |
0.89 |
| chr9_12775321_12775833 | 0.29 |
LURAP1L |
leucine rich adaptor protein 1-like |
557 |
0.78 |
| chr15_84116498_84116804 | 0.29 |
SH3GL3 |
SH3-domain GRB2-like 3 |
368 |
0.92 |
| chr16_69998972_69999655 | 0.29 |
CLEC18A |
C-type lectin domain family 18, member A |
13669 |
0.14 |
| chr22_30796002_30796153 | 0.29 |
SEC14L2 |
SEC14-like 2 (S. cerevisiae) |
1511 |
0.18 |
| chr2_177001412_177001990 | 0.29 |
HOXD3 |
homeobox D3 |
16 |
0.5 |
| chr17_2310404_2311021 | 0.28 |
MNT |
MAX network transcriptional repressor |
6300 |
0.12 |
| chr10_62760865_62761099 | 0.28 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
216 |
0.97 |
| chr1_31887122_31887273 | 0.28 |
SERINC2 |
serine incorporator 2 |
537 |
0.74 |
| chr13_107185066_107185426 | 0.28 |
EFNB2 |
ephrin-B2 |
2216 |
0.4 |
| chr2_165477169_165477439 | 0.27 |
GRB14 |
growth factor receptor-bound protein 14 |
684 |
0.79 |
| chr19_56166514_56167932 | 0.27 |
U2AF2 |
U2 small nuclear RNA auxiliary factor 2 |
792 |
0.34 |
| chr3_184300809_184300975 | 0.27 |
EPHB3 |
EPH receptor B3 |
21320 |
0.18 |
| chr17_75474006_75474173 | 0.27 |
SEPT9 |
septin 9 |
2764 |
0.22 |
| chr1_183236957_183237240 | 0.27 |
NMNAT2 |
nicotinamide nucleotide adenylyltransferase 2 |
36911 |
0.19 |
| chr11_61486524_61486808 | 0.26 |
MYRF |
myelin regulatory factor |
33455 |
0.11 |
| chr7_27225817_27225968 | 0.26 |
HOXA11-AS |
HOXA11 antisense RNA |
739 |
0.31 |
| chr4_151503456_151503607 | 0.26 |
MAB21L2 |
mab-21-like 2 (C. elegans) |
454 |
0.84 |
| chr10_88728248_88728709 | 0.26 |
ADIRF |
adipogenesis regulatory factor |
289 |
0.5 |
| chr20_23031599_23032105 | 0.25 |
THBD |
thrombomodulin |
1474 |
0.34 |
| chr9_140506939_140507169 | 0.25 |
C9orf37 |
chromosome 9 open reading frame 37 |
6211 |
0.12 |
| chr8_128864051_128864244 | 0.25 |
ENSG00000249859 |
. |
55939 |
0.13 |
| chr14_69015576_69015827 | 0.25 |
CTD-2325P2.4 |
|
79461 |
0.1 |
| chr2_75787048_75787447 | 0.25 |
EVA1A |
eva-1 homolog A (C. elegans) |
792 |
0.61 |
| chr8_99014292_99014443 | 0.25 |
MATN2 |
matrilin 2 |
14405 |
0.15 |
| chr17_79438730_79438973 | 0.25 |
RP11-1055B8.8 |
|
9888 |
0.1 |
| chr10_103330110_103330718 | 0.24 |
DPCD |
deleted in primary ciliary dyskinesia homolog (mouse) |
8 |
0.98 |
| chr5_112582093_112582348 | 0.24 |
MCC |
mutated in colorectal cancers |
11846 |
0.27 |
| chr19_5578062_5578357 | 0.24 |
SAFB2 |
scaffold attachment factor B2 |
44782 |
0.11 |
| chr2_127643626_127643777 | 0.24 |
AC114783.1 |
Protein LOC339760 |
12771 |
0.28 |
| chr3_71158955_71159120 | 0.24 |
FOXP1 |
forkhead box P1 |
20707 |
0.28 |
| chr9_81871858_81872685 | 0.24 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
314417 |
0.01 |
| chr1_32263785_32263971 | 0.24 |
SPOCD1 |
SPOC domain containing 1 |
439 |
0.79 |
| chr11_58730865_58731226 | 0.24 |
RP11-142C4.6 |
|
15713 |
0.15 |
| chr16_65155038_65155536 | 0.23 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
546 |
0.88 |
| chr3_114169976_114170457 | 0.23 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
3314 |
0.33 |
| chr6_85484117_85484534 | 0.23 |
TBX18 |
T-box 18 |
10088 |
0.3 |
| chr15_96100470_96100831 | 0.23 |
ENSG00000222076 |
. |
188383 |
0.03 |
| chrX_34675593_34676078 | 0.23 |
TMEM47 |
transmembrane protein 47 |
430 |
0.92 |
| chr17_45197869_45198099 | 0.23 |
ENSG00000221016 |
. |
2116 |
0.25 |
| chr22_46933754_46934840 | 0.22 |
CELSR1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
1230 |
0.46 |
| chr15_75944753_75944922 | 0.22 |
SNX33 |
sorting nexin 33 |
2740 |
0.14 |
| chr12_124004050_124004201 | 0.22 |
RILPL1 |
Rab interacting lysosomal protein-like 1 |
5150 |
0.17 |
| chr2_202723535_202724043 | 0.22 |
ENSG00000221374 |
. |
17937 |
0.16 |
| chr12_120032319_120032470 | 0.22 |
TMEM233 |
transmembrane protein 233 |
1130 |
0.49 |
| chr2_133173748_133173945 | 0.22 |
GPR39 |
G protein-coupled receptor 39 |
301 |
0.92 |
| chr8_42063953_42065062 | 0.22 |
PLAT |
plasminogen activator, tissue |
576 |
0.71 |
| chr9_134287538_134287689 | 0.21 |
PRRC2B |
proline-rich coiled-coil 2B |
17864 |
0.19 |
| chr11_89597318_89597927 | 0.21 |
TRIM64B |
tripartite motif containing 64B |
11563 |
0.22 |
| chr7_131240036_131240570 | 0.21 |
PODXL |
podocalyxin-like |
1063 |
0.65 |
| chr4_164265165_164265569 | 0.21 |
NPY5R |
neuropeptide Y receptor Y5 |
113 |
0.9 |
| chr7_28997325_28997548 | 0.21 |
CTB-113D17.1 |
|
22147 |
0.2 |
| chr2_151343410_151343893 | 0.21 |
RND3 |
Rho family GTPase 3 |
530 |
0.88 |
| chr9_116356336_116356765 | 0.21 |
RGS3 |
regulator of G-protein signaling 3 |
784 |
0.67 |
| chr20_43970794_43971114 | 0.21 |
SDC4 |
syndecan 4 |
6110 |
0.13 |
| chr8_120879463_120879614 | 0.21 |
ENSG00000244642 |
. |
4636 |
0.17 |
| chr14_58332211_58332467 | 0.21 |
SLC35F4 |
solute carrier family 35, member F4 |
441 |
0.87 |
| chr3_157155512_157155783 | 0.21 |
PTX3 |
pentraxin 3, long |
1069 |
0.63 |
| chr3_66007529_66007703 | 0.20 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
16367 |
0.19 |
| chr21_47394558_47394771 | 0.20 |
COL6A1 |
collagen, type VI, alpha 1 |
6987 |
0.21 |
| chr11_1896066_1896383 | 0.20 |
LSP1 |
lymphocyte-specific protein 1 |
1483 |
0.24 |
| chr16_2085550_2085759 | 0.20 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
2382 |
0.09 |
| chr7_64035084_64035528 | 0.20 |
ZNF680 |
zinc finger protein 680 |
11822 |
0.28 |
| chr1_2569102_2569253 | 0.20 |
MMEL1 |
membrane metallo-endopeptidase-like 1 |
4696 |
0.16 |
| chr15_68573802_68573977 | 0.20 |
FEM1B |
fem-1 homolog b (C. elegans) |
1592 |
0.39 |
| chr17_77997988_77998139 | 0.20 |
CTD-2529O21.2 |
|
333 |
0.85 |
| chr2_16832297_16832583 | 0.20 |
FAM49A |
family with sequence similarity 49, member A |
14656 |
0.3 |
| chr17_6922316_6922467 | 0.20 |
RP11-589P10.7 |
|
342 |
0.53 |
| chr18_57386327_57386692 | 0.20 |
CCBE1 |
collagen and calcium binding EGF domains 1 |
21897 |
0.15 |
| chr3_149094469_149095370 | 0.20 |
TM4SF1-AS1 |
TM4SF1 antisense RNA 1 |
646 |
0.47 |
| chr8_22443451_22443742 | 0.19 |
AC037459.4 |
Uncharacterized protein |
3191 |
0.12 |
| chr12_26266071_26266589 | 0.19 |
SSPN |
sarcospan |
8594 |
0.18 |
| chr19_51455841_51455992 | 0.19 |
KLK5 |
kallikrein-related peptidase 5 |
282 |
0.77 |
| chr1_204042818_204043418 | 0.19 |
SOX13 |
SRY (sex determining region Y)-box 13 |
36 |
0.98 |
| chr6_4733420_4733615 | 0.19 |
CDYL |
chromodomain protein, Y-like |
27124 |
0.2 |
| chr20_48599552_48599945 | 0.19 |
SNAI1 |
snail family zinc finger 1 |
212 |
0.92 |
| chr12_96183320_96183471 | 0.19 |
NTN4 |
netrin 4 |
331 |
0.86 |
| chr2_36678228_36678401 | 0.19 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
62519 |
0.12 |
| chr19_41195400_41196566 | 0.19 |
NUMBL |
numb homolog (Drosophila)-like |
44 |
0.96 |
| chr4_57203544_57203695 | 0.19 |
ENSG00000251823 |
. |
6236 |
0.18 |
| chr19_7953457_7953661 | 0.19 |
LRRC8E |
leucine rich repeat containing 8 family, member E |
123 |
0.88 |
| chr19_13983804_13984015 | 0.19 |
ENSG00000207613 |
. |
1604 |
0.17 |
| chr13_110979643_110979794 | 0.19 |
COL4A2 |
collagen, type IV, alpha 2 |
20104 |
0.2 |
| chr3_44040787_44041403 | 0.18 |
ENSG00000252980 |
. |
71484 |
0.12 |
| chr10_70359131_70359282 | 0.18 |
TET1 |
tet methylcytosine dioxygenase 1 |
38793 |
0.13 |
| chr17_75523143_75523294 | 0.18 |
SEPT9 |
septin 9 |
45223 |
0.13 |
| chr7_130131272_130131861 | 0.18 |
MEST |
mesoderm specific transcript |
368 |
0.77 |
| chr15_68580618_68580911 | 0.18 |
FEM1B |
fem-1 homolog b (C. elegans) |
829 |
0.64 |
| chr1_6556688_6556983 | 0.18 |
PLEKHG5 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
79 |
0.95 |
| chr11_833873_834149 | 0.18 |
CD151 |
CD151 molecule (Raph blood group) |
861 |
0.26 |
| chr9_110513848_110514093 | 0.18 |
AL162389.1 |
Uncharacterized protein |
26449 |
0.19 |
| chr17_75404849_75405000 | 0.18 |
SEPT9 |
septin 9 |
3765 |
0.19 |
| chr20_3657828_3657979 | 0.18 |
ADAM33 |
ADAM metallopeptidase domain 33 |
4847 |
0.17 |
| chr9_137445335_137445486 | 0.18 |
COL5A1 |
collagen, type V, alpha 1 |
88210 |
0.08 |
| chr20_43729394_43729952 | 0.18 |
KCNS1 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
77 |
0.96 |
| chr17_77752893_77753171 | 0.18 |
CBX2 |
chromobox homolog 2 |
1039 |
0.43 |
| chr12_47471040_47471191 | 0.17 |
PCED1B |
PC-esterase domain containing 1B |
2271 |
0.31 |
| chr1_150480831_150481087 | 0.17 |
ECM1 |
extracellular matrix protein 1 |
376 |
0.77 |
| chr6_28641788_28642196 | 0.17 |
ENSG00000272278 |
. |
25950 |
0.18 |
| chr11_61159610_61160825 | 0.17 |
TMEM216 |
transmembrane protein 216 |
352 |
0.79 |
| chr16_70221940_70222878 | 0.17 |
CLEC18C |
C-type lectin domain family 18, member C |
14349 |
0.1 |
| chr9_137221469_137221673 | 0.17 |
RXRA |
retinoid X receptor, alpha |
3145 |
0.33 |
| chr5_140753330_140753954 | 0.17 |
PCDHGA6 |
protocadherin gamma subfamily A, 6 |
9 |
0.91 |
| chr12_6930752_6931082 | 0.17 |
GPR162 |
G protein-coupled receptor 162 |
47 |
0.93 |
| chr15_74344226_74344377 | 0.17 |
ENSG00000244612 |
. |
20738 |
0.13 |
| chr17_77925559_77925739 | 0.17 |
TBC1D16 |
TBC1 domain family, member 16 |
163 |
0.96 |
| chr20_25565265_25566139 | 0.17 |
NINL |
ninein-like |
451 |
0.81 |
| chr12_132316928_132317093 | 0.17 |
MMP17 |
matrix metallopeptidase 17 (membrane-inserted) |
3925 |
0.2 |
| chr17_74542570_74542721 | 0.17 |
RP11-666A8.7 |
|
3388 |
0.1 |
| chr16_51188825_51189552 | 0.17 |
SALL1 |
spalt-like transcription factor 1 |
4002 |
0.28 |
| chr11_43955270_43955437 | 0.17 |
C11orf96 |
chromosome 11 open reading frame 96 |
8461 |
0.16 |
| chr1_39569795_39569946 | 0.16 |
MACF1 |
microtubule-actin crosslinking factor 1 |
1213 |
0.46 |
| chr17_41909774_41910015 | 0.16 |
MPP3 |
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
284 |
0.86 |
| chr17_36902558_36902709 | 0.16 |
PCGF2 |
polycomb group ring finger 2 |
248 |
0.78 |
| chr1_67142291_67142442 | 0.16 |
AL139147.1 |
Uncharacterized protein |
344 |
0.88 |
| chr22_32022509_32022660 | 0.16 |
PISD |
phosphatidylserine decarboxylase |
304 |
0.91 |
| chr20_10634629_10634907 | 0.16 |
JAG1 |
jagged 1 |
8386 |
0.25 |
| chr1_45082713_45082906 | 0.16 |
RNF220 |
ring finger protein 220 |
9189 |
0.17 |
| chr7_73245792_73245943 | 0.16 |
CLDN4 |
claudin 4 |
674 |
0.63 |
| chr15_40616235_40616987 | 0.16 |
LINC00984 |
long intergenic non-protein coding RNA 984 |
806 |
0.39 |
| chr16_4348992_4349159 | 0.16 |
GLIS2 |
GLIS family zinc finger 2 |
15687 |
0.13 |
| chr22_21311135_21311286 | 0.16 |
ENSG00000266366 |
. |
1761 |
0.2 |
| chr9_695532_695683 | 0.16 |
RP11-130C19.3 |
|
10052 |
0.22 |
| chr14_100041328_100041479 | 0.16 |
CCDC85C |
coiled-coil domain containing 85C |
1526 |
0.37 |
| chr16_74440490_74441404 | 0.16 |
CLEC18B |
C-type lectin domain family 18, member B |
14343 |
0.14 |
| chr5_73770430_73770700 | 0.16 |
ENSG00000244326 |
. |
156754 |
0.04 |
| chr7_5458775_5459294 | 0.15 |
AC093620.5 |
|
424 |
0.46 |
| chr16_31548902_31549248 | 0.15 |
AHSP |
alpha hemoglobin stabilizing protein |
9777 |
0.11 |
| chr3_48777290_48777441 | 0.15 |
PRKAR2A |
protein kinase, cAMP-dependent, regulatory, type II, alpha |
12386 |
0.13 |
| chr3_175244326_175245006 | 0.15 |
ENSG00000201648 |
. |
82601 |
0.1 |
| chr11_393450_393735 | 0.15 |
PKP3 |
plakophilin 3 |
129 |
0.92 |
| chr4_125825441_125825711 | 0.15 |
ANKRD50 |
ankyrin repeat domain 50 |
191689 |
0.03 |
| chr17_68165729_68165994 | 0.15 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
185 |
0.66 |
| chr2_58655964_58656121 | 0.15 |
FANCL |
Fanconi anemia, complementation group L |
187535 |
0.03 |
| chr5_140762477_140762699 | 0.15 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
121 |
0.87 |
| chr8_27491561_27492240 | 0.15 |
SCARA3 |
scavenger receptor class A, member 3 |
202 |
0.93 |
| chr1_40421563_40422081 | 0.15 |
MFSD2A |
major facilitator superfamily domain containing 2A |
1000 |
0.48 |
| chr22_19946584_19946980 | 0.15 |
COMT |
catechol-O-methyltransferase |
3288 |
0.15 |
| chr6_21886388_21886539 | 0.15 |
ENSG00000222515 |
. |
199310 |
0.03 |
| chr11_63803897_63804159 | 0.15 |
RP11-21A7A.2 |
|
586 |
0.65 |
| chr2_151335102_151335779 | 0.15 |
RND3 |
Rho family GTPase 3 |
6456 |
0.35 |
| chr6_80656707_80657785 | 0.15 |
ELOVL4 |
ELOVL fatty acid elongase 4 |
51 |
0.98 |
| chr3_133967997_133968801 | 0.15 |
RYK |
receptor-like tyrosine kinase |
1038 |
0.66 |
| chr1_150480597_150480804 | 0.15 |
ECM1 |
extracellular matrix protein 1 |
117 |
0.94 |
| chrX_22153526_22153707 | 0.15 |
PHEX-AS1 |
PHEX antisense RNA 1 |
37484 |
0.16 |
| chr3_138957437_138957611 | 0.14 |
MRPS22 |
mitochondrial ribosomal protein S22 |
105337 |
0.06 |
| chr8_131503545_131503696 | 0.14 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
47714 |
0.2 |
| chr13_110959773_110960124 | 0.14 |
COL4A2 |
collagen, type IV, alpha 2 |
334 |
0.6 |
| chr5_14036568_14037099 | 0.14 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
92181 |
0.09 |
| chr6_56716060_56716427 | 0.14 |
DST |
dystonin |
506 |
0.82 |
| chr14_23237291_23237442 | 0.14 |
OXA1L |
oxidase (cytochrome c) assembly 1-like |
1438 |
0.23 |
| chr1_156718751_156718902 | 0.14 |
HDGF |
hepatoma-derived growth factor |
2259 |
0.16 |
| chr20_4129930_4130735 | 0.14 |
SMOX |
spermine oxidase |
750 |
0.71 |
| chr1_223921059_223921210 | 0.14 |
CAPN2 |
calpain 2, (m/II) large subunit |
21100 |
0.19 |
| chr14_55121110_55121261 | 0.14 |
SAMD4A |
sterile alpha motif domain containing 4A |
86548 |
0.09 |
| chr16_4424352_4424847 | 0.14 |
VASN |
vasorin |
2750 |
0.17 |
| chr8_54789629_54790008 | 0.14 |
RP11-1070A24.2 |
|
609 |
0.71 |
| chr13_36705915_36706066 | 0.14 |
DCLK1 |
doublecortin-like kinase 1 |
547 |
0.84 |
| chr3_122786767_122787219 | 0.14 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
798 |
0.71 |
| chr1_209603784_209603935 | 0.14 |
ENSG00000230937 |
. |
1619 |
0.52 |
| chr19_47163250_47163584 | 0.14 |
DACT3-AS1 |
DACT3 antisense RNA 1 |
204 |
0.83 |
| chrX_153601896_153602918 | 0.14 |
FLNA |
filamin A, alpha |
587 |
0.54 |
| chr13_72440002_72440456 | 0.14 |
DACH1 |
dachshund homolog 1 (Drosophila) |
678 |
0.82 |
| chr13_110972530_110972681 | 0.14 |
COL4A2 |
collagen, type IV, alpha 2 |
12991 |
0.21 |
| chr12_109901272_109901510 | 0.14 |
KCTD10 |
potassium channel tetramerization domain containing 10 |
3191 |
0.21 |
| chr14_78447028_78447687 | 0.14 |
ENSG00000199440 |
. |
159872 |
0.03 |
| chr11_45922009_45922186 | 0.14 |
MAPK8IP1 |
mitogen-activated protein kinase 8 interacting protein 1 |
3996 |
0.14 |
| chr2_236578616_236578898 | 0.14 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
509 |
0.87 |
| chr17_77767116_77767731 | 0.14 |
CBX8 |
chromobox homolog 8 |
3492 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.4 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.1 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.3 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.2 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.2 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.2 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.0 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.0 | 0.2 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.0 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.1 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.0 | GO:0060453 | regulation of gastric acid secretion(GO:0060453) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.2 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0006853 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.5 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.0 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | Genes involved in Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |