Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
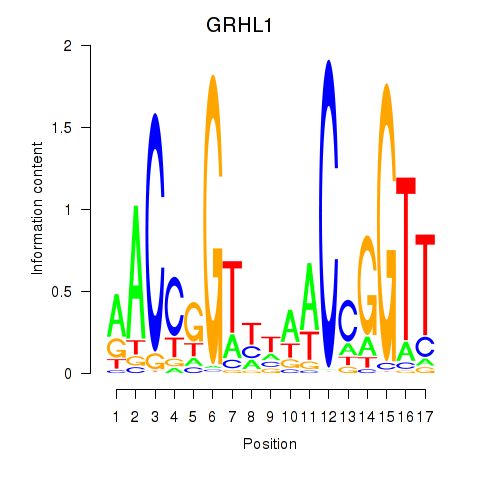
Results for GRHL1
Z-value: 1.36
Transcription factors associated with GRHL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GRHL1
|
ENSG00000134317.13 | grainyhead like transcription factor 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_10091040_10091475 | GRHL1 | 535 | 0.791189 | 0.91 | 6.9e-04 | Click! |
| chr2_10090883_10091034 | GRHL1 | 834 | 0.639056 | 0.82 | 6.2e-03 | Click! |
| chr2_10090679_10090830 | GRHL1 | 1038 | 0.553917 | 0.81 | 8.5e-03 | Click! |
| chr2_10091778_10091990 | GRHL1 | 57 | 0.977081 | 0.77 | 1.6e-02 | Click! |
| chr2_10124648_10124876 | GRHL1 | 10672 | 0.165414 | 0.51 | 1.6e-01 | Click! |
Activity of the GRHL1 motif across conditions
Conditions sorted by the z-value of the GRHL1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_34626880_34627184 | 0.70 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
2013 |
0.2 |
| chr2_10091778_10091990 | 0.62 |
GRHL1 |
grainyhead-like 1 (Drosophila) |
57 |
0.98 |
| chr1_147142364_147142605 | 0.62 |
ACP6 |
acid phosphatase 6, lysophosphatidic |
120 |
0.97 |
| chr8_81262205_81262356 | 0.58 |
ENSG00000206649 |
. |
32976 |
0.18 |
| chr21_46493485_46493673 | 0.58 |
AP001579.1 |
Uncharacterized protein |
652 |
0.5 |
| chr11_104838069_104839428 | 0.57 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
1345 |
0.45 |
| chr10_31321494_31321708 | 0.56 |
ZNF438 |
zinc finger protein 438 |
735 |
0.75 |
| chr6_108497000_108497194 | 0.55 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
7130 |
0.18 |
| chr14_99734743_99735157 | 0.54 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
2615 |
0.3 |
| chr7_100487833_100488038 | 0.54 |
UFSP1 |
UFM1-specific peptidase 1 (non-functional) |
596 |
0.54 |
| chr9_140121732_140121883 | 0.53 |
RNF224 |
ring finger protein 224 |
211 |
0.78 |
| chr19_1491435_1491715 | 0.51 |
REEP6 |
receptor accessory protein 6 |
410 |
0.58 |
| chr16_433636_433787 | 0.51 |
TMEM8A |
transmembrane protein 8A |
1729 |
0.2 |
| chr3_128444656_128444807 | 0.51 |
RAB7A |
RAB7A, member RAS oncogene family |
234 |
0.92 |
| chr7_69946018_69946169 | 0.47 |
AUTS2 |
autism susceptibility candidate 2 |
248032 |
0.02 |
| chr17_71087973_71088223 | 0.47 |
SLC39A11 |
solute carrier family 39, member 11 |
717 |
0.75 |
| chr1_206732914_206733127 | 0.47 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
2527 |
0.25 |
| chr6_159084795_159085026 | 0.47 |
SYTL3 |
synaptotagmin-like 3 |
715 |
0.68 |
| chr8_24151584_24151953 | 0.46 |
ADAM28 |
ADAM metallopeptidase domain 28 |
144 |
0.97 |
| chr16_72236428_72236579 | 0.46 |
PMFBP1 |
polyamine modulated factor 1 binding protein 1 |
25726 |
0.19 |
| chr5_126147731_126147991 | 0.46 |
LMNB1 |
lamin B1 |
34980 |
0.19 |
| chr5_141016577_141016949 | 0.45 |
RELL2 |
RELT-like 2 |
170 |
0.67 |
| chr8_28558312_28558856 | 0.43 |
EXTL3-AS1 |
EXTL3 antisense RNA 1 |
186 |
0.6 |
| chrX_119602028_119603217 | 0.43 |
LAMP2 |
lysosomal-associated membrane protein 2 |
225 |
0.95 |
| chr3_172240144_172241280 | 0.42 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
553 |
0.83 |
| chr1_44016013_44016164 | 0.42 |
PTPRF |
protein tyrosine phosphatase, receptor type, F |
5341 |
0.22 |
| chr17_77788750_77788949 | 0.41 |
ENSG00000238331 |
. |
4371 |
0.16 |
| chr12_54376113_54376277 | 0.41 |
HOXC-AS3 |
HOXC cluster antisense RNA 3 |
1913 |
0.11 |
| chr7_39989218_39989369 | 0.41 |
RP11-467D6.1 |
|
61 |
0.83 |
| chr16_1582253_1582404 | 0.40 |
IFT140 |
intraflagellar transport 140 homolog (Chlamydomonas) |
1167 |
0.25 |
| chr2_158297089_158297418 | 0.40 |
ENSG00000216054 |
. |
809 |
0.56 |
| chr9_4662590_4663079 | 0.40 |
PPAPDC2 |
phosphatidic acid phosphatase type 2 domain containing 2 |
536 |
0.74 |
| chr4_89534455_89534711 | 0.39 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
7638 |
0.19 |
| chr3_182986311_182986462 | 0.38 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
3254 |
0.26 |
| chr1_150976711_150976969 | 0.38 |
FAM63A |
family with sequence similarity 63, member A |
2121 |
0.14 |
| chr8_38215915_38216066 | 0.38 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
22349 |
0.11 |
| chr9_133031265_133031416 | 0.37 |
HMCN2 |
hemicentin 2 |
15542 |
0.23 |
| chr2_52954585_52954736 | 0.37 |
ENSG00000264975 |
. |
24907 |
0.29 |
| chr7_100183497_100183687 | 0.37 |
LRCH4 |
leucine-rich repeats and calponin homology (CH) domain containing 4 |
184 |
0.64 |
| chr18_39534653_39534946 | 0.36 |
PIK3C3 |
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
372 |
0.93 |
| chr1_29066884_29067035 | 0.36 |
YTHDF2 |
YTH domain family, member 2 |
3482 |
0.22 |
| chr1_12240704_12240855 | 0.36 |
ENSG00000263676 |
. |
10991 |
0.15 |
| chr22_20424024_20424175 | 0.36 |
ENSG00000271796 |
. |
10054 |
0.12 |
| chr5_173665504_173665655 | 0.35 |
CTC-229L21.1 |
|
65475 |
0.14 |
| chr17_18265908_18266237 | 0.35 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
588 |
0.63 |
| chrX_97323503_97323654 | 0.34 |
DIAPH2-AS1 |
DIAPH2 antisense RNA 1 |
425990 |
0.01 |
| chr21_35305474_35305968 | 0.34 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
2203 |
0.25 |
| chr17_55434039_55434190 | 0.33 |
ENSG00000263902 |
. |
55448 |
0.13 |
| chr13_111364022_111364765 | 0.33 |
ING1 |
inhibitor of growth family, member 1 |
690 |
0.72 |
| chr2_113592071_113592291 | 0.33 |
IL1B |
interleukin 1, beta |
1830 |
0.31 |
| chr19_40030594_40030834 | 0.33 |
EID2 |
EP300 interacting inhibitor of differentiation 2 |
156 |
0.91 |
| chr8_142231340_142231491 | 0.33 |
SLC45A4 |
solute carrier family 45, member 4 |
7254 |
0.16 |
| chr4_154388914_154389185 | 0.33 |
KIAA0922 |
KIAA0922 |
1548 |
0.49 |
| chr1_25060751_25060902 | 0.32 |
CLIC4 |
chloride intracellular channel 4 |
11022 |
0.2 |
| chr11_62572291_62572857 | 0.32 |
NXF1 |
nuclear RNA export factor 1 |
91 |
0.89 |
| chr13_41540610_41540946 | 0.32 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
15640 |
0.18 |
| chr18_29021056_29021207 | 0.32 |
DSG3 |
desmoglein 3 |
6627 |
0.17 |
| chr22_21775304_21775479 | 0.31 |
HIC2 |
hypermethylated in cancer 2 |
466 |
0.71 |
| chr2_85640695_85641186 | 0.31 |
CAPG |
capping protein (actin filament), gelsolin-like |
222 |
0.85 |
| chr8_96150063_96150319 | 0.31 |
PLEKHF2 |
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
3887 |
0.25 |
| chr12_48099956_48100625 | 0.31 |
RPAP3 |
RNA polymerase II associated protein 3 |
446 |
0.81 |
| chr16_73031152_73031303 | 0.31 |
ENSG00000221799 |
. |
12471 |
0.2 |
| chr9_37030099_37030250 | 0.31 |
PAX5 |
paired box 5 |
3854 |
0.25 |
| chr1_159870035_159870397 | 0.31 |
CCDC19 |
coiled-coil domain containing 19 |
263 |
0.55 |
| chr12_120667761_120668330 | 0.31 |
PXN |
paxillin |
3398 |
0.15 |
| chr3_149956404_149956555 | 0.31 |
RP11-167H9.4 |
|
140660 |
0.04 |
| chr6_116797520_116797797 | 0.31 |
RP1-93H18.6 |
|
12803 |
0.11 |
| chr16_4818256_4818411 | 0.31 |
ZNF500 |
zinc finger protein 500 |
708 |
0.52 |
| chr18_9018040_9018191 | 0.31 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
84513 |
0.08 |
| chr2_109236287_109236438 | 0.30 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
1360 |
0.51 |
| chr22_36769485_36769706 | 0.30 |
MYH9 |
myosin, heavy chain 9, non-muscle |
8438 |
0.19 |
| chr13_106996229_106996778 | 0.30 |
ENSG00000222682 |
. |
188664 |
0.03 |
| chr3_69139558_69139806 | 0.30 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
5510 |
0.16 |
| chr10_75815554_75815957 | 0.30 |
VCL |
vinculin |
27479 |
0.17 |
| chr6_35265630_35266434 | 0.30 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
403 |
0.84 |
| chr7_151548013_151548400 | 0.30 |
PRKAG2-AS1 |
PRKAG2 antisense RNA 1 |
25921 |
0.14 |
| chr12_30848459_30848714 | 0.30 |
IPO8 |
importin 8 |
334 |
0.92 |
| chr2_241832903_241833054 | 0.30 |
C2orf54 |
chromosome 2 open reading frame 54 |
1523 |
0.34 |
| chr12_64798382_64799443 | 0.29 |
XPOT |
exportin, tRNA |
86 |
0.97 |
| chr14_68431244_68431431 | 0.29 |
RAD51B |
RAD51 paralog B |
141076 |
0.04 |
| chr18_72166510_72167447 | 0.29 |
CNDP2 |
CNDP dipeptidase 2 (metallopeptidase M20 family) |
118 |
0.97 |
| chr11_46141542_46141818 | 0.29 |
PHF21A |
PHD finger protein 21A |
267 |
0.92 |
| chr17_7834732_7835241 | 0.29 |
TRAPPC1 |
trafficking protein particle complex 1 |
277 |
0.58 |
| chr8_17293592_17293743 | 0.29 |
MTMR7 |
myotubularin related protein 7 |
22831 |
0.22 |
| chr12_102454924_102455862 | 0.29 |
CCDC53 |
coiled-coil domain containing 53 |
464 |
0.76 |
| chr1_51426093_51426543 | 0.29 |
CDKN2C |
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
99 |
0.82 |
| chr19_1795974_1796208 | 0.29 |
ATP8B3 |
ATPase, aminophospholipid transporter, class I, type 8B, member 3 |
15532 |
0.09 |
| chr18_12839173_12839633 | 0.28 |
PTPN2 |
protein tyrosine phosphatase, non-receptor type 2 |
2580 |
0.33 |
| chr2_69004074_69004225 | 0.28 |
ARHGAP25 |
Rho GTPase activating protein 25 |
2077 |
0.39 |
| chr20_57431129_57431280 | 0.28 |
GNAS |
GNAS complex locus |
971 |
0.45 |
| chr18_45834501_45834652 | 0.28 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
30186 |
0.19 |
| chr13_98084501_98085278 | 0.27 |
RAP2A |
RAP2A, member of RAS oncogene family |
1587 |
0.5 |
| chr6_25963703_25964110 | 0.27 |
TRIM38 |
tripartite motif containing 38 |
631 |
0.58 |
| chr17_58228097_58228436 | 0.27 |
CA4 |
carbonic anhydrase IV |
963 |
0.55 |
| chr12_120907252_120907411 | 0.27 |
SRSF9 |
serine/arginine-rich splicing factor 9 |
43 |
0.8 |
| chr19_15435123_15435274 | 0.27 |
BRD4 |
bromodomain containing 4 |
7519 |
0.18 |
| chr9_131534376_131534797 | 0.27 |
ZER1 |
zyg-11 related, cell cycle regulator |
107 |
0.94 |
| chr5_112257522_112257903 | 0.26 |
REEP5 |
receptor accessory protein 5 |
18 |
0.97 |
| chr9_95570304_95570464 | 0.26 |
ANKRD19P |
ankyrin repeat domain 19, pseudogene |
1509 |
0.37 |
| chr2_201991504_201992128 | 0.26 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
2236 |
0.22 |
| chr6_24666286_24667027 | 0.26 |
TDP2 |
tyrosyl-DNA phosphodiesterase 2 |
163 |
0.84 |
| chr16_23568244_23568679 | 0.26 |
UBFD1 |
ubiquitin family domain containing 1 |
69 |
0.68 |
| chr18_44496350_44497016 | 0.26 |
PIAS2 |
protein inhibitor of activated STAT, 2 |
359 |
0.71 |
| chr12_106751930_106752216 | 0.25 |
POLR3B |
polymerase (RNA) III (DNA directed) polypeptide B |
213 |
0.95 |
| chr22_21274799_21275119 | 0.25 |
CRKL |
v-crk avian sarcoma virus CT10 oncogene homolog-like |
3245 |
0.15 |
| chr20_45986315_45986612 | 0.25 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
896 |
0.5 |
| chrX_2815751_2815902 | 0.25 |
ARSD-AS1 |
ARSD antisense RNA 1 |
7119 |
0.17 |
| chr17_47335011_47335162 | 0.25 |
PHOSPHO1 |
phosphatase, orphan 1 |
26958 |
0.13 |
| chr14_22192285_22192436 | 0.25 |
ENSG00000222776 |
. |
56425 |
0.12 |
| chr19_13209170_13209477 | 0.25 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
4358 |
0.12 |
| chr20_56861857_56862008 | 0.25 |
PPP4R1L |
protein phosphatase 4, regulatory subunit 1-like |
22563 |
0.18 |
| chr19_40596081_40597156 | 0.25 |
AC005614.3 |
|
43 |
0.64 |
| chrX_111214548_111214699 | 0.25 |
TRPC5OS |
TRPC5 opposite strand |
89498 |
0.09 |
| chrX_40027620_40027771 | 0.25 |
BCOR |
BCL6 corepressor |
8878 |
0.31 |
| chr16_74481592_74481743 | 0.24 |
ENSG00000251794 |
. |
8772 |
0.15 |
| chr14_51403011_51403162 | 0.24 |
PYGL |
phosphorylase, glycogen, liver |
8088 |
0.19 |
| chr5_134094525_134094724 | 0.24 |
DDX46 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
130 |
0.95 |
| chr6_16130421_16130915 | 0.24 |
MYLIP |
myosin regulatory light chain interacting protein |
1266 |
0.45 |
| chr10_50626942_50627093 | 0.24 |
RP11-123B3.2 |
|
306 |
0.9 |
| chr12_54018173_54018459 | 0.24 |
ATF7 |
activating transcription factor 7 |
1439 |
0.29 |
| chr22_46436516_46436667 | 0.24 |
RP6-109B7.5 |
|
12382 |
0.1 |
| chr19_34663821_34664709 | 0.24 |
LSM14A |
LSM14A, SCD6 homolog A (S. cerevisiae) |
702 |
0.77 |
| chr1_149711963_149712172 | 0.24 |
ENSG00000263825 |
. |
3116 |
0.14 |
| chr5_176786759_176786924 | 0.24 |
RGS14 |
regulator of G-protein signaling 14 |
2003 |
0.18 |
| chr3_182513059_182513572 | 0.23 |
ATP11B |
ATPase, class VI, type 11B |
2024 |
0.32 |
| chr16_23782392_23782543 | 0.23 |
CHP2 |
calcineurin-like EF-hand protein 2 |
16519 |
0.17 |
| chr16_3414793_3414984 | 0.23 |
MTRNR2L4 |
MT-RNR2-like 4 |
7395 |
0.11 |
| chr9_132514519_132514800 | 0.23 |
PTGES |
prostaglandin E synthase |
667 |
0.66 |
| chr1_193154702_193155811 | 0.23 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
528 |
0.8 |
| chr10_55884978_55885129 | 0.23 |
ENSG00000263921 |
. |
222721 |
0.02 |
| chr2_213880427_213880578 | 0.22 |
AC079610.1 |
|
5767 |
0.31 |
| chr8_42748827_42749465 | 0.22 |
ENSG00000266044 |
. |
2272 |
0.19 |
| chr9_77641918_77642364 | 0.22 |
C9orf41 |
chromosome 9 open reading frame 41 |
126 |
0.96 |
| chr11_113433650_113433801 | 0.22 |
DRD2 |
dopamine receptor D2 |
87614 |
0.08 |
| chr13_20805734_20806347 | 0.22 |
GJB6 |
gap junction protein, beta 6, 30kDa |
400 |
0.87 |
| chr12_1313659_1314218 | 0.22 |
ERC1 |
ELKS/RAB6-interacting/CAST family member 1 |
177000 |
0.03 |
| chr12_15039325_15039476 | 0.22 |
MGP |
matrix Gla protein |
540 |
0.72 |
| chr1_6532288_6532439 | 0.22 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
6108 |
0.12 |
| chr1_151030627_151031763 | 0.22 |
CDC42SE1 |
CDC42 small effector 1 |
930 |
0.27 |
| chr2_191183369_191183553 | 0.22 |
HIBCH |
3-hydroxyisobutyryl-CoA hydrolase |
1155 |
0.54 |
| chr17_18091438_18091835 | 0.22 |
RP11-258F1.1 |
|
3569 |
0.14 |
| chr1_114888713_114889187 | 0.21 |
TRIM33 |
tripartite motif containing 33 |
116908 |
0.06 |
| chr8_71126242_71126393 | 0.21 |
NCOA2 |
nuclear receptor coactivator 2 |
31293 |
0.22 |
| chr14_59655511_59655710 | 0.21 |
DAAM1 |
dishevelled associated activator of morphogenesis 1 |
193 |
0.97 |
| chr10_64134972_64135123 | 0.21 |
ZNF365 |
zinc finger protein 365 |
256 |
0.94 |
| chr1_38454967_38455422 | 0.21 |
SF3A3 |
splicing factor 3a, subunit 3, 60kDa |
465 |
0.7 |
| chr19_6669496_6669799 | 0.21 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
481 |
0.75 |
| chr16_58843484_58843635 | 0.21 |
ENSG00000244003 |
. |
39131 |
0.16 |
| chr16_49561945_49562197 | 0.21 |
ZNF423 |
zinc finger protein 423 |
112692 |
0.06 |
| chr17_4711251_4711402 | 0.21 |
PLD2 |
phospholipase D2 |
469 |
0.63 |
| chr1_70670699_70671189 | 0.21 |
LRRC40 |
leucine rich repeat containing 40 |
359 |
0.53 |
| chrX_11777788_11778628 | 0.21 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
461 |
0.9 |
| chr3_16883621_16883873 | 0.21 |
PLCL2 |
phospholipase C-like 2 |
42705 |
0.19 |
| chr10_99168347_99168498 | 0.21 |
RRP12 |
ribosomal RNA processing 12 homolog (S. cerevisiae) |
7295 |
0.1 |
| chr6_28907953_28908476 | 0.21 |
C6orf100 |
chromosome 6 open reading frame 100 |
3440 |
0.16 |
| chrX_52004796_52004996 | 0.21 |
RP11-363G10.2 |
|
61933 |
0.11 |
| chr19_2042439_2042863 | 0.20 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
19 |
0.96 |
| chr9_125011063_125011347 | 0.20 |
ENSG00000272317 |
. |
7007 |
0.13 |
| chr1_249152113_249152387 | 0.20 |
ZNF692 |
zinc finger protein 692 |
809 |
0.43 |
| chr14_70161423_70161574 | 0.20 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
32119 |
0.19 |
| chr11_72492321_72492601 | 0.20 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
419 |
0.73 |
| chr2_37880790_37881166 | 0.20 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
6177 |
0.28 |
| chrX_70329371_70329522 | 0.20 |
IL2RG |
interleukin 2 receptor, gamma |
328 |
0.79 |
| chr1_75196025_75196269 | 0.20 |
CRYZ |
crystallin, zeta (quinone reductase) |
2348 |
0.29 |
| chr1_68215148_68215597 | 0.20 |
ENSG00000238778 |
. |
22964 |
0.2 |
| chr17_143390_143541 | 0.20 |
RP11-1260E13.1 |
|
27718 |
0.13 |
| chr17_33412938_33413089 | 0.20 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
2841 |
0.14 |
| chr1_210406047_210406239 | 0.20 |
SERTAD4 |
SERTA domain containing 4 |
1 |
0.69 |
| chr2_101729475_101729669 | 0.20 |
ENSG00000265860 |
. |
19541 |
0.19 |
| chr7_134832844_134833492 | 0.20 |
AC083862.1 |
Uncharacterized protein |
252 |
0.58 |
| chr4_107227768_107227919 | 0.20 |
TBCK |
TBC1 domain containing kinase |
3902 |
0.28 |
| chr9_77703527_77704915 | 0.20 |
OSTF1 |
osteoclast stimulating factor 1 |
762 |
0.5 |
| chrX_65217747_65217898 | 0.20 |
RP6-159A1.3 |
|
1771 |
0.4 |
| chr2_102314830_102315631 | 0.20 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
238 |
0.96 |
| chr6_47009797_47009948 | 0.20 |
GPR110 |
G protein-coupled receptor 110 |
158 |
0.97 |
| chr10_52176688_52176970 | 0.20 |
AC069547.2 |
Uncharacterized protein |
24063 |
0.19 |
| chr10_75011688_75012124 | 0.19 |
MRPS16 |
mitochondrial ribosomal protein S16 |
484 |
0.47 |
| chr16_2588059_2588223 | 0.19 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
24 |
0.92 |
| chr9_5450042_5450535 | 0.19 |
CD274 |
CD274 molecule |
215 |
0.94 |
| chr5_176800994_176801145 | 0.19 |
SLC34A1 |
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
5167 |
0.1 |
| chr15_71740308_71740459 | 0.19 |
ENSG00000252813 |
. |
27443 |
0.23 |
| chr15_66679290_66679512 | 0.19 |
MAP2K1 |
mitogen-activated protein kinase kinase 1 |
246 |
0.56 |
| chr11_26604890_26605185 | 0.19 |
MUC15 |
mucin 15, cell surface associated |
11257 |
0.3 |
| chr19_3163912_3164063 | 0.19 |
AC005264.2 |
|
8814 |
0.11 |
| chr15_40366035_40366186 | 0.19 |
RP11-521C20.3 |
|
14923 |
0.14 |
| chr22_43330177_43330519 | 0.19 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
11531 |
0.23 |
| chr6_153323257_153323895 | 0.19 |
MTRF1L |
mitochondrial translational release factor 1-like |
244 |
0.91 |
| chr9_84301810_84302157 | 0.19 |
TLE1 |
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
300 |
0.93 |
| chr19_53700941_53701337 | 0.19 |
ZNF665 |
zinc finger protein 665 |
4512 |
0.14 |
| chr10_126410360_126410511 | 0.19 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
17682 |
0.16 |
| chr2_74433191_74433522 | 0.18 |
MTHFD2 |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
7613 |
0.14 |
| chr17_46048416_46049087 | 0.18 |
CDK5RAP3 |
CDK5 regulatory subunit associated protein 3 |
234 |
0.85 |
| chr2_46542537_46542756 | 0.18 |
EPAS1 |
endothelial PAS domain protein 1 |
18105 |
0.25 |
| chr17_53829632_53829783 | 0.18 |
PCTP |
phosphatidylcholine transfer protein |
946 |
0.66 |
| chr12_88429328_88429956 | 0.18 |
C12orf29 |
chromosome 12 open reading frame 29 |
419 |
0.84 |
| chr11_77113473_77113624 | 0.18 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
9401 |
0.24 |
| chr15_82648106_82648257 | 0.18 |
GOLGA6L10 |
golgin A6 family-like 10 |
6475 |
0.18 |
| chr12_120912385_120912536 | 0.18 |
DYNLL1 |
dynein, light chain, LC8-type 1 |
4774 |
0.11 |
| chr12_52611181_52611332 | 0.18 |
KRT7 |
keratin 7 |
15642 |
0.1 |
| chr8_29206404_29207399 | 0.18 |
DUSP4 |
dual specificity phosphatase 4 |
579 |
0.71 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.2 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 0.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.2 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.1 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.2 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.1 | 0.2 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:1903312 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.5 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.1 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.2 | GO:0006385 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.3 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.0 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.2 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.4 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:1901224 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.0 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.2 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.0 | GO:0097035 | regulation of membrane lipid distribution(GO:0097035) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.1 | GO:0043032 | positive regulation of macrophage activation(GO:0043032) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0042130 | negative regulation of T cell proliferation(GO:0042130) |
| 0.0 | 0.0 | GO:0002335 | mature B cell differentiation involved in immune response(GO:0002313) mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0046719 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:0000080 | mitotic G1 phase(GO:0000080) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0060346 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.0 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.2 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.1 | 0.2 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.1 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.4 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.0 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0046970 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.5 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |