Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
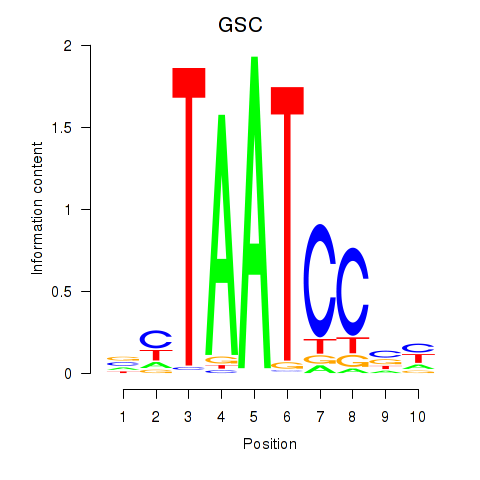
Results for GSC_GSC2
Z-value: 0.75
Transcription factors associated with GSC_GSC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GSC
|
ENSG00000133937.3 | goosecoid homeobox |
|
GSC2
|
ENSG00000063515.2 | goosecoid homeobox 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_95363694_95363845 | GSC | 127207 | 0.051511 | -0.85 | 3.8e-03 | Click! |
| chr14_95218837_95218988 | GSC | 17650 | 0.246162 | -0.59 | 9.4e-02 | Click! |
| chr14_95242002_95242153 | GSC | 5515 | 0.300144 | -0.52 | 1.6e-01 | Click! |
| chr14_95402811_95402962 | GSC | 166324 | 0.032050 | -0.45 | 2.2e-01 | Click! |
| chr14_95236319_95236561 | GSC | 122 | 0.978358 | 0.41 | 2.8e-01 | Click! |
| chr22_19136719_19137042 | GSC2 | 916 | 0.414342 | 0.81 | 7.8e-03 | Click! |
| chr22_19137054_19137376 | GSC2 | 581 | 0.606263 | 0.68 | 4.4e-02 | Click! |
| chr22_19138892_19139181 | GSC2 | 1240 | 0.310193 | 0.43 | 2.5e-01 | Click! |
Activity of the GSC_GSC2 motif across conditions
Conditions sorted by the z-value of the GSC_GSC2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
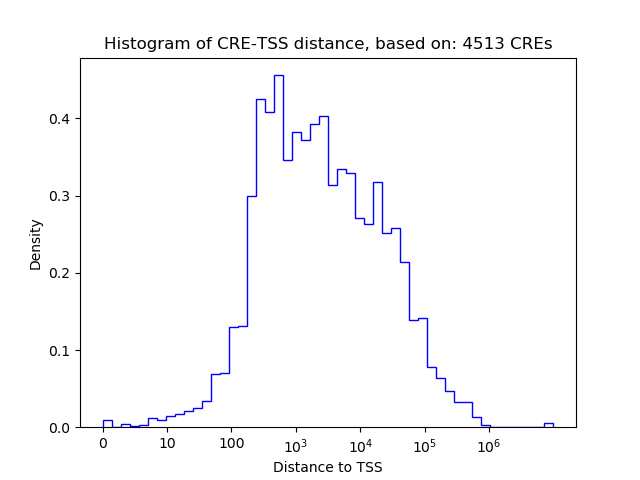
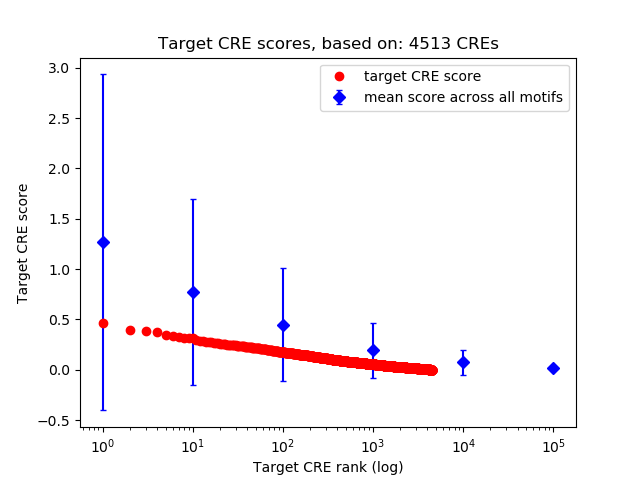
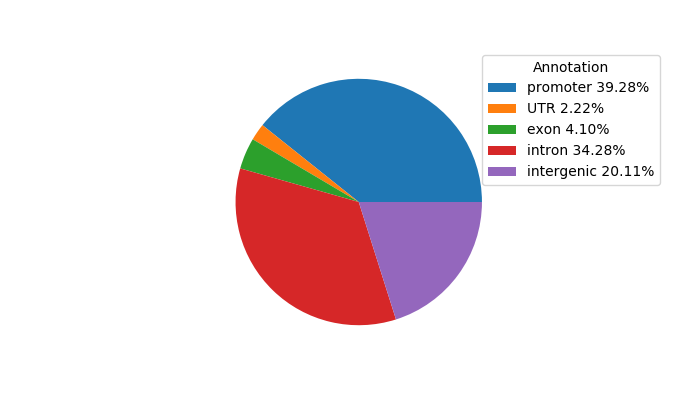
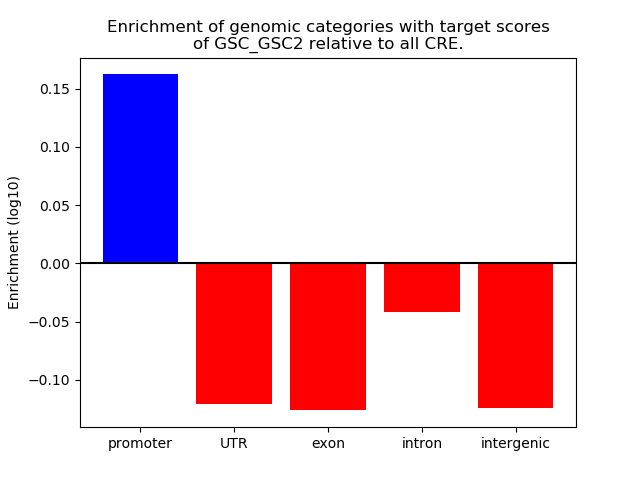
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_128383033_128383215 | 0.46 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
7835 |
0.24 |
| chr4_109083741_109084005 | 0.40 |
LEF1 |
lymphoid enhancer-binding factor 1 |
3584 |
0.27 |
| chr17_73522364_73522644 | 0.39 |
LLGL2 |
lethal giant larvae homolog 2 (Drosophila) |
634 |
0.56 |
| chr1_198610531_198610895 | 0.38 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
2421 |
0.36 |
| chr19_48752565_48752795 | 0.34 |
CARD8 |
caspase recruitment domain family, member 8 |
245 |
0.88 |
| chr16_27415626_27415898 | 0.34 |
IL21R |
interleukin 21 receptor |
1339 |
0.47 |
| chr12_122729504_122729668 | 0.33 |
VPS33A |
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
5126 |
0.17 |
| chr12_47606378_47606570 | 0.32 |
PCED1B |
PC-esterase domain containing 1B |
3578 |
0.27 |
| chr20_43597121_43597440 | 0.31 |
STK4 |
serine/threonine kinase 4 |
2113 |
0.25 |
| chr1_226922484_226922931 | 0.31 |
ITPKB |
inositol-trisphosphate 3-kinase B |
2452 |
0.34 |
| chr11_66314663_66314814 | 0.30 |
ZDHHC24 |
zinc finger, DHHC-type containing 24 |
1029 |
0.34 |
| chr14_22952184_22952433 | 0.29 |
ENSG00000251002 |
. |
360 |
0.73 |
| chr4_40200858_40201138 | 0.28 |
RHOH |
ras homolog family member H |
966 |
0.6 |
| chr2_113931904_113932161 | 0.28 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
458 |
0.63 |
| chr3_16329620_16329771 | 0.27 |
OXNAD1 |
oxidoreductase NAD-binding domain containing 1 |
18947 |
0.15 |
| chr12_51713121_51713481 | 0.27 |
BIN2 |
bridging integrator 2 |
4598 |
0.18 |
| chr2_192540821_192541028 | 0.27 |
NABP1 |
nucleic acid binding protein 1 |
1938 |
0.47 |
| chr12_109254375_109254526 | 0.27 |
DAO |
D-amino-acid oxidase |
1742 |
0.29 |
| chrX_21676275_21676631 | 0.26 |
KLHL34 |
kelch-like family member 34 |
5 |
0.99 |
| chr3_60066334_60066485 | 0.26 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
108826 |
0.08 |
| chrX_12991784_12992037 | 0.26 |
TMSB4X |
thymosin beta 4, X-linked |
1317 |
0.51 |
| chr19_6425433_6425584 | 0.26 |
KHSRP |
KH-type splicing regulatory protein |
703 |
0.46 |
| chr12_4381475_4382675 | 0.25 |
CCND2 |
cyclin D2 |
863 |
0.52 |
| chr1_25889390_25889576 | 0.25 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
19412 |
0.19 |
| chr4_3304601_3305007 | 0.25 |
RGS12 |
regulator of G-protein signaling 12 |
5958 |
0.26 |
| chr9_469822_470055 | 0.25 |
RP11-165F24.3 |
|
102 |
0.79 |
| chr5_134879321_134879990 | 0.24 |
NEUROG1 |
neurogenin 1 |
8016 |
0.18 |
| chr14_22466666_22466817 | 0.24 |
ENSG00000238634 |
. |
144146 |
0.04 |
| chr12_103889282_103889433 | 0.24 |
C12orf42 |
chromosome 12 open reading frame 42 |
374 |
0.91 |
| chr3_63953694_63954206 | 0.24 |
ATXN7 |
ataxin 7 |
530 |
0.74 |
| chr13_41372571_41372761 | 0.24 |
SLC25A15 |
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 |
5372 |
0.18 |
| chr11_104904746_104905919 | 0.24 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
508 |
0.77 |
| chr22_40304150_40304385 | 0.24 |
GRAP2 |
GRB2-related adaptor protein 2 |
7154 |
0.18 |
| chr14_100544647_100544798 | 0.23 |
CTD-2376I20.1 |
|
3507 |
0.18 |
| chrX_48771508_48771659 | 0.23 |
PIM2 |
pim-2 oncogene |
1429 |
0.23 |
| chr13_68203141_68203292 | 0.23 |
PCDH9 |
protocadherin 9 |
398748 |
0.01 |
| chr15_40599321_40599893 | 0.23 |
PLCB2 |
phospholipase C, beta 2 |
419 |
0.68 |
| chr19_35630816_35630967 | 0.23 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
35 |
0.95 |
| chrX_11780001_11780185 | 0.23 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
2346 |
0.44 |
| chr6_32160504_32160787 | 0.23 |
GPSM3 |
G-protein signaling modulator 3 |
0 |
0.94 |
| chr20_34331618_34331769 | 0.23 |
RBM39 |
RNA binding motif protein 39 |
1459 |
0.29 |
| chr5_141062081_141062481 | 0.23 |
ARAP3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
493 |
0.75 |
| chr2_175459196_175459684 | 0.22 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
3053 |
0.24 |
| chr1_117299964_117300188 | 0.22 |
CD2 |
CD2 molecule |
2987 |
0.29 |
| chr14_22204676_22204903 | 0.22 |
ENSG00000222776 |
. |
43996 |
0.15 |
| chr13_40532306_40532606 | 0.22 |
ENSG00000212553 |
. |
101092 |
0.08 |
| chr3_112217855_112218341 | 0.22 |
BTLA |
B and T lymphocyte associated |
107 |
0.98 |
| chr6_128239129_128239316 | 0.22 |
THEMIS |
thymocyte selection associated |
463 |
0.89 |
| chr4_26886197_26886348 | 0.22 |
STIM2 |
stromal interaction molecule 2 |
23191 |
0.22 |
| chr17_38019635_38019786 | 0.22 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
669 |
0.62 |
| chr20_42360174_42360325 | 0.22 |
GTSF1L |
gametocyte specific factor 1-like |
4611 |
0.26 |
| chr2_198173822_198174153 | 0.22 |
ANKRD44 |
ankyrin repeat domain 44 |
1524 |
0.32 |
| chr4_81106153_81106599 | 0.21 |
PRDM8 |
PR domain containing 8 |
48 |
0.98 |
| chr15_80262648_80263745 | 0.21 |
BCL2A1 |
BCL2-related protein A1 |
315 |
0.89 |
| chr2_27487055_27487432 | 0.21 |
SLC30A3 |
solute carrier family 30 (zinc transporter), member 3 |
239 |
0.85 |
| chr16_3627461_3627612 | 0.21 |
NLRC3 |
NLR family, CARD domain containing 3 |
135 |
0.95 |
| chr5_159713959_159714110 | 0.21 |
ENSG00000243654 |
. |
4761 |
0.2 |
| chr3_113250860_113251011 | 0.21 |
SIDT1 |
SID1 transmembrane family, member 1 |
208 |
0.94 |
| chr18_56326442_56326593 | 0.21 |
MALT1 |
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
12101 |
0.14 |
| chr20_61438769_61438978 | 0.21 |
OGFR |
opioid growth factor receptor |
248 |
0.85 |
| chr10_70362033_70362257 | 0.21 |
TET1 |
tet methylcytosine dioxygenase 1 |
41732 |
0.13 |
| chr15_100890052_100890414 | 0.21 |
ADAMTS17 |
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
8023 |
0.23 |
| chr8_134509504_134509655 | 0.20 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
2025 |
0.48 |
| chr14_22203899_22204050 | 0.20 |
ENSG00000222776 |
. |
44811 |
0.15 |
| chr16_67970526_67970748 | 0.20 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
353 |
0.73 |
| chr6_106969857_106970345 | 0.20 |
AIM1 |
absent in melanoma 1 |
10371 |
0.2 |
| chr19_53496276_53496464 | 0.20 |
ERVV-1 |
endogenous retrovirus group V, member 1 |
20804 |
0.13 |
| chr16_85769625_85769776 | 0.20 |
ENSG00000222190 |
. |
5606 |
0.13 |
| chr10_14612285_14612533 | 0.20 |
FAM107B |
family with sequence similarity 107, member B |
1620 |
0.47 |
| chr14_22989269_22989534 | 0.20 |
TRAJ15 |
T cell receptor alpha joining 15 |
9179 |
0.11 |
| chr6_154568354_154568864 | 0.19 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
53 |
0.99 |
| chr14_22739869_22740020 | 0.19 |
ENSG00000238634 |
. |
129057 |
0.04 |
| chr4_69433676_69433827 | 0.19 |
UGT2B17 |
UDP glucuronosyltransferase 2 family, polypeptide B17 |
494 |
0.87 |
| chr14_94543541_94543837 | 0.19 |
DDX24 |
DEAD (Asp-Glu-Ala-Asp) box helicase 24 |
3859 |
0.17 |
| chr14_61814123_61814302 | 0.19 |
PRKCH |
protein kinase C, eta |
397 |
0.87 |
| chr7_36327271_36327770 | 0.19 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
9233 |
0.18 |
| chr21_35306376_35306527 | 0.19 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
2933 |
0.21 |
| chr5_40684385_40684698 | 0.19 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
4941 |
0.23 |
| chr8_100714756_100715301 | 0.19 |
ENSG00000243254 |
. |
88722 |
0.08 |
| chr4_149367147_149367363 | 0.19 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
1405 |
0.59 |
| chr1_25260510_25260661 | 0.19 |
RUNX3 |
runt-related transcription factor 3 |
4217 |
0.26 |
| chr1_66800579_66801197 | 0.19 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
3016 |
0.39 |
| chr4_40240088_40240239 | 0.18 |
RHOH |
ras homolog family member H |
38199 |
0.16 |
| chr10_120891057_120891670 | 0.18 |
FAM45A |
family with sequence similarity 45, member A |
27734 |
0.11 |
| chr6_37104092_37104243 | 0.18 |
PIM1 |
pim-1 oncogene |
33812 |
0.15 |
| chr6_2902163_2902328 | 0.18 |
SERPINB9 |
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
1269 |
0.44 |
| chr3_72788121_72788360 | 0.18 |
ENSG00000222838 |
. |
47278 |
0.17 |
| chr1_155146016_155146243 | 0.18 |
TRIM46 |
tripartite motif containing 46 |
225 |
0.54 |
| chr11_108099098_108099249 | 0.18 |
ENSG00000206967 |
. |
1158 |
0.42 |
| chr8_65607910_65608061 | 0.18 |
RP11-1D12.1 |
|
39031 |
0.2 |
| chr2_238581678_238581829 | 0.18 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
19035 |
0.2 |
| chr1_202612976_202613127 | 0.18 |
SYT2 |
synaptotagmin II |
470 |
0.84 |
| chr5_118623311_118623849 | 0.18 |
ENSG00000243333 |
. |
18746 |
0.17 |
| chr13_99951513_99951834 | 0.18 |
GPR183 |
G protein-coupled receptor 183 |
7986 |
0.2 |
| chrX_44203680_44203969 | 0.18 |
EFHC2 |
EF-hand domain (C-terminal) containing 2 |
906 |
0.71 |
| chr11_133814931_133815363 | 0.18 |
IGSF9B |
immunoglobulin superfamily, member 9B |
1040 |
0.55 |
| chr6_139461625_139461834 | 0.18 |
HECA |
headcase homolog (Drosophila) |
5480 |
0.28 |
| chr7_38315629_38315780 | 0.18 |
STARD3NL |
STARD3 N-terminal like |
97707 |
0.09 |
| chr11_104787850_104788088 | 0.18 |
RP11-693N9.2 |
|
7327 |
0.2 |
| chr9_132402475_132402626 | 0.18 |
RP11-483H20.4 |
|
205 |
0.89 |
| chr1_154324818_154325020 | 0.18 |
ENSG00000238365 |
. |
13700 |
0.1 |
| chr2_240188602_240189192 | 0.17 |
ENSG00000265215 |
. |
38260 |
0.14 |
| chr22_51059908_51060313 | 0.17 |
ARSA |
arylsulfatase A |
6473 |
0.1 |
| chr11_128372497_128372648 | 0.17 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
2717 |
0.34 |
| chr14_23015065_23015417 | 0.17 |
AE000662.92 |
Uncharacterized protein |
10293 |
0.11 |
| chr1_66799510_66799957 | 0.17 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
1861 |
0.5 |
| chr7_150416321_150416472 | 0.17 |
GIMAP1 |
GTPase, IMAP family member 1 |
2751 |
0.23 |
| chr3_187726002_187726153 | 0.17 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
144995 |
0.04 |
| chr20_52226068_52226301 | 0.17 |
ZNF217 |
zinc finger protein 217 |
753 |
0.69 |
| chr6_30648112_30648263 | 0.17 |
PPP1R18 |
protein phosphatase 1, regulatory subunit 18 |
6806 |
0.07 |
| chr19_9902856_9903204 | 0.17 |
ZNF846 |
zinc finger protein 846 |
743 |
0.48 |
| chr3_16342795_16343107 | 0.17 |
RP11-415F23.2 |
|
12995 |
0.16 |
| chr2_169347644_169347797 | 0.17 |
CERS6 |
ceramide synthase 6 |
34961 |
0.16 |
| chr1_198620819_198621442 | 0.17 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
12838 |
0.23 |
| chr2_219082127_219082488 | 0.17 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
374 |
0.79 |
| chr6_143256936_143257087 | 0.17 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
9327 |
0.26 |
| chr9_37036362_37036543 | 0.17 |
PAX5 |
paired box 5 |
2349 |
0.32 |
| chr1_116918127_116918427 | 0.17 |
ATP1A1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
1788 |
0.31 |
| chr1_12675825_12676469 | 0.17 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
1590 |
0.37 |
| chr3_151911560_151911983 | 0.17 |
MBNL1 |
muscleblind-like splicing regulator 1 |
74058 |
0.11 |
| chr18_18685478_18685632 | 0.17 |
ROCK1 |
Rho-associated, coiled-coil containing protein kinase 1 |
6257 |
0.22 |
| chr1_154379631_154380335 | 0.17 |
RP11-350G8.5 |
|
943 |
0.42 |
| chr15_45930000_45930287 | 0.16 |
SQRDL |
sulfide quinone reductase-like (yeast) |
3205 |
0.24 |
| chr7_151572690_151573009 | 0.16 |
PRKAG2-AS1 |
PRKAG2 antisense RNA 1 |
1278 |
0.31 |
| chr12_6445436_6445689 | 0.16 |
TNFRSF1A |
tumor necrosis factor receptor superfamily, member 1A |
5452 |
0.13 |
| chr16_2265337_2265543 | 0.16 |
PGP |
phosphoglycolate phosphatase |
632 |
0.39 |
| chr1_198614847_198615101 | 0.16 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
6682 |
0.25 |
| chr7_23338458_23338640 | 0.16 |
MALSU1 |
mitochondrial assembly of ribosomal large subunit 1 |
270 |
0.93 |
| chr2_201989949_201990411 | 0.16 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
2980 |
0.18 |
| chr8_27235851_27236002 | 0.16 |
PTK2B |
protein tyrosine kinase 2 beta |
2242 |
0.36 |
| chr1_156784010_156784282 | 0.16 |
NTRK1 |
neurotrophic tyrosine kinase, receptor, type 1 |
1302 |
0.3 |
| chr11_65409143_65409294 | 0.16 |
SIPA1 |
signal-induced proliferation-associated 1 |
919 |
0.33 |
| chr22_31139652_31139831 | 0.16 |
ENSG00000264661 |
. |
12197 |
0.17 |
| chr2_162849236_162849504 | 0.16 |
ENSG00000253046 |
. |
74078 |
0.1 |
| chrY_14773573_14773724 | 0.16 |
USP9Y |
ubiquitin specific peptidase 9, Y-linked |
39512 |
0.22 |
| chr7_129546585_129546878 | 0.16 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
44436 |
0.1 |
| chrX_129217430_129217581 | 0.16 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
26831 |
0.18 |
| chr19_39825870_39826313 | 0.16 |
GMFG |
glia maturation factor, gamma |
554 |
0.58 |
| chr7_143106132_143106395 | 0.16 |
EPHA1 |
EPH receptor A1 |
278 |
0.82 |
| chr2_233926827_233927087 | 0.16 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
1768 |
0.35 |
| chr19_4302092_4302295 | 0.16 |
TMIGD2 |
transmembrane and immunoglobulin domain containing 2 |
189 |
0.87 |
| chr18_2641583_2641787 | 0.16 |
ENSG00000200875 |
. |
8181 |
0.15 |
| chr7_142334440_142334591 | 0.16 |
MTRNR2L6 |
MT-RNR2-like 6 |
39589 |
0.18 |
| chr17_28706240_28706541 | 0.16 |
CPD |
carboxypeptidase D |
467 |
0.76 |
| chrX_80353779_80353930 | 0.16 |
HMGN5 |
high mobility group nucleosome binding domain 5 |
23283 |
0.25 |
| chr2_201676360_201676545 | 0.16 |
BZW1 |
basic leucine zipper and W2 domains 1 |
183 |
0.73 |
| chr7_129420780_129420931 | 0.15 |
ENSG00000207691 |
. |
6001 |
0.16 |
| chr2_73181298_73181449 | 0.15 |
EMX1 |
empty spiracles homeobox 1 |
30256 |
0.19 |
| chr16_79472215_79472382 | 0.15 |
MAF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
161501 |
0.04 |
| chr22_38791219_38791718 | 0.15 |
CSNK1E |
casein kinase 1, epsilon |
3059 |
0.17 |
| chr16_30887671_30887822 | 0.15 |
MIR4519 |
microRNA 4519 |
513 |
0.52 |
| chr1_198652510_198652661 | 0.15 |
RP11-553K8.5 |
|
16395 |
0.24 |
| chr17_7239932_7240900 | 0.15 |
ACAP1 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
507 |
0.51 |
| chr11_2198846_2198997 | 0.15 |
ENSG00000265258 |
. |
4628 |
0.12 |
| chr8_23075544_23076103 | 0.15 |
ENSG00000246582 |
. |
6161 |
0.12 |
| chrX_30473325_30473476 | 0.15 |
CXorf21 |
chromosome X open reading frame 21 |
122561 |
0.05 |
| chr2_143889174_143889389 | 0.15 |
ARHGAP15 |
Rho GTPase activating protein 15 |
2398 |
0.38 |
| chr19_4068105_4068310 | 0.15 |
ZBTB7A |
zinc finger and BTB domain containing 7A |
1264 |
0.32 |
| chr16_85768387_85768538 | 0.15 |
ENSG00000222190 |
. |
6844 |
0.12 |
| chr15_34620156_34620427 | 0.15 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
8754 |
0.11 |
| chr6_2900436_2900587 | 0.15 |
SERPINB9 |
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
3003 |
0.24 |
| chr6_32810383_32810534 | 0.15 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
1339 |
0.24 |
| chr12_21606326_21606605 | 0.15 |
PYROXD1 |
pyridine nucleotide-disulphide oxidoreductase domain 1 |
15616 |
0.17 |
| chr17_56419518_56419935 | 0.15 |
BZRAP1-AS1 |
BZRAP1 antisense RNA 1 |
5007 |
0.12 |
| chr16_68269729_68269880 | 0.15 |
ESRP2 |
epithelial splicing regulatory protein 2 |
231 |
0.8 |
| chr1_206827719_206827870 | 0.15 |
DYRK3 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
18620 |
0.12 |
| chr12_55525482_55525633 | 0.15 |
OR9K2 |
olfactory receptor, family 9, subfamily K, member 2 |
2092 |
0.37 |
| chr10_76345165_76345892 | 0.15 |
ENSG00000206756 |
. |
34378 |
0.2 |
| chr21_25801249_25801648 | 0.15 |
ENSG00000232512 |
. |
671997 |
0.0 |
| chr19_48759351_48759594 | 0.15 |
CARD8 |
caspase recruitment domain family, member 8 |
269 |
0.69 |
| chr2_237446743_237446920 | 0.15 |
ACKR3 |
atypical chemokine receptor 3 |
29599 |
0.19 |
| chr11_46848529_46848759 | 0.15 |
CKAP5 |
cytoskeleton associated protein 5 |
240 |
0.91 |
| chrX_111922694_111923357 | 0.15 |
LHFPL1 |
lipoma HMGIC fusion partner-like 1 |
257 |
0.94 |
| chr1_38471684_38471863 | 0.15 |
FHL3 |
four and a half LIM domains 3 |
596 |
0.64 |
| chr17_5001058_5001209 | 0.15 |
ENSG00000234327 |
. |
13630 |
0.09 |
| chr5_146257138_146257401 | 0.15 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
936 |
0.67 |
| chr10_90760175_90760505 | 0.15 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
9193 |
0.15 |
| chr20_52225525_52225734 | 0.14 |
ZNF217 |
zinc finger protein 217 |
198 |
0.95 |
| chr11_118036813_118036964 | 0.14 |
SCN2B |
sodium channel, voltage-gated, type II, beta subunit |
10500 |
0.14 |
| chr4_54232426_54232649 | 0.14 |
SCFD2 |
sec1 family domain containing 2 |
295 |
0.92 |
| chr11_2323518_2323777 | 0.14 |
TSPAN32 |
tetraspanin 32 |
267 |
0.57 |
| chr4_90228457_90229530 | 0.14 |
GPRIN3 |
GPRIN family member 3 |
168 |
0.97 |
| chr12_15107546_15107731 | 0.14 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
3553 |
0.2 |
| chr17_21155590_21155741 | 0.14 |
C17orf103 |
chromosome 17 open reading frame 103 |
913 |
0.52 |
| chr11_63719409_63719560 | 0.14 |
NAA40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
12764 |
0.1 |
| chr19_48614158_48614373 | 0.14 |
PLA2G4C |
phospholipase A2, group IVC (cytosolic, calcium-independent) |
191 |
0.91 |
| chr7_104908860_104909472 | 0.14 |
SRPK2 |
SRSF protein kinase 2 |
296 |
0.92 |
| chr5_55775454_55775711 | 0.14 |
CTC-236F12.4 |
Uncharacterized protein |
2014 |
0.36 |
| chr16_11676228_11676379 | 0.14 |
LITAF |
lipopolysaccharide-induced TNF factor |
3926 |
0.23 |
| chr18_56105832_56106078 | 0.14 |
ENSG00000207778 |
. |
12351 |
0.17 |
| chr6_130070208_130070770 | 0.14 |
ENSG00000211996 |
. |
1607 |
0.48 |
| chr1_226068766_226068917 | 0.14 |
TMEM63A |
transmembrane protein 63A |
1228 |
0.35 |
| chr17_39741665_39741816 | 0.14 |
KRT14 |
keratin 14 |
1433 |
0.25 |
| chr6_17704634_17704930 | 0.14 |
RP11-500C11.3 |
|
1706 |
0.27 |
| chr15_91475141_91475394 | 0.14 |
HDDC3 |
HD domain containing 3 |
439 |
0.62 |
| chr14_23013027_23013178 | 0.14 |
AE000662.92 |
Uncharacterized protein |
12432 |
0.1 |
| chr1_111745814_111746254 | 0.14 |
DENND2D |
DENN/MADD domain containing 2D |
997 |
0.41 |
| chr18_11907803_11908245 | 0.14 |
MPPE1 |
metallophosphoesterase 1 |
248 |
0.78 |
| chr22_30720802_30721186 | 0.14 |
TBC1D10A |
TBC1 domain family, member 10A |
1896 |
0.18 |
| chr18_74836762_74837008 | 0.14 |
MBP |
myelin basic protein |
2783 |
0.4 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.3 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.1 | 0.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.4 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.2 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0002669 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.3 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0010536 | regulation of activation of Janus kinase activity(GO:0010533) positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.2 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0032367 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.0 | GO:1903514 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 | 0.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0046794 | multi-organism transport(GO:0044766) transport of virus(GO:0046794) intracellular transport of virus(GO:0075733) multi-organism localization(GO:1902579) multi-organism intracellular transport(GO:1902583) |
| 0.0 | 0.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.0 | 0.0 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0045978 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.0 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.0 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.2 | GO:0030101 | natural killer cell activation(GO:0030101) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.0 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.0 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.0 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.0 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |