Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
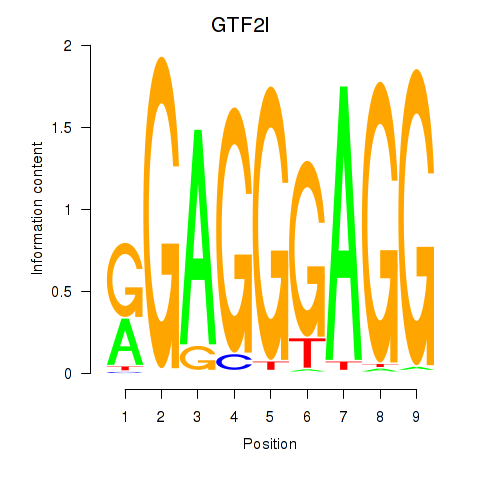
Results for GTF2I
Z-value: 0.49
Transcription factors associated with GTF2I
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GTF2I
|
ENSG00000077809.8 | general transcription factor IIi |
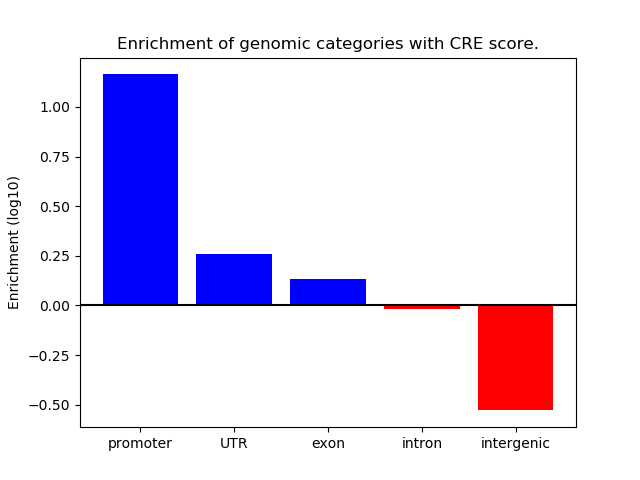
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_74042267_74042418 | GTF2I | 29669 | 0.173457 | 0.83 | 5.3e-03 | Click! |
| chr7_74077316_74077467 | GTF2I | 5034 | 0.251743 | 0.78 | 1.3e-02 | Click! |
| chr7_74087036_74087187 | GTF2I | 11880 | 0.216977 | 0.66 | 5.4e-02 | Click! |
| chr7_74075100_74075450 | GTF2I | 2918 | 0.305488 | 0.63 | 6.6e-02 | Click! |
| chr7_74074778_74074955 | GTF2I | 2509 | 0.330487 | 0.47 | 2.1e-01 | Click! |
Activity of the GTF2I motif across conditions
Conditions sorted by the z-value of the GTF2I motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
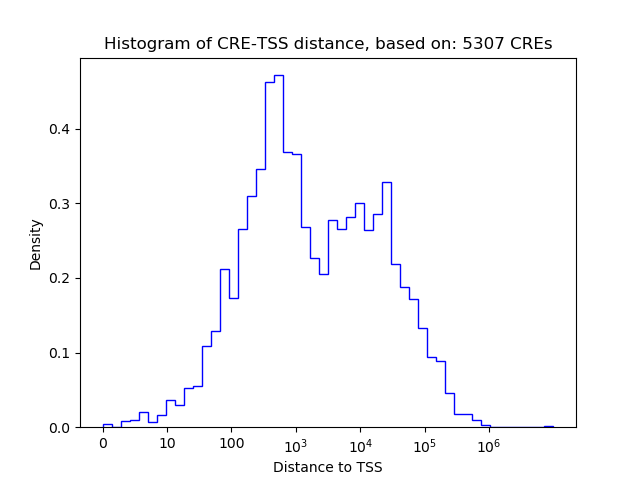
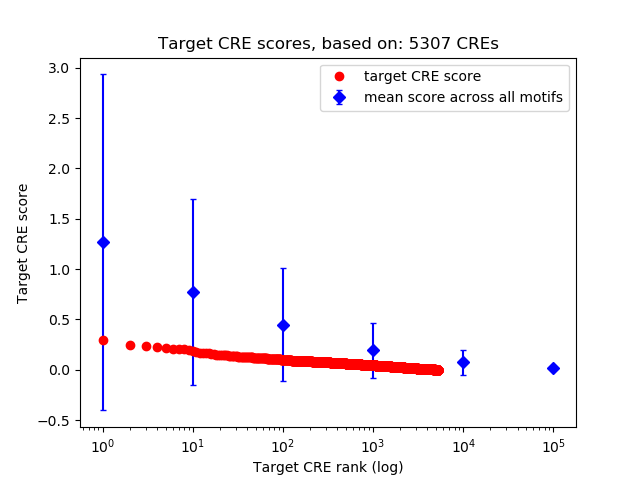
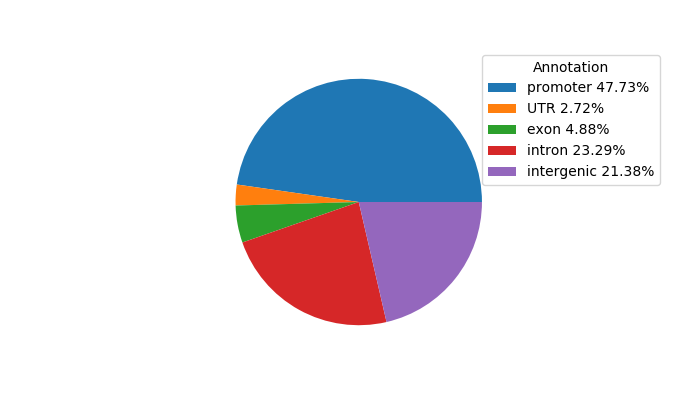
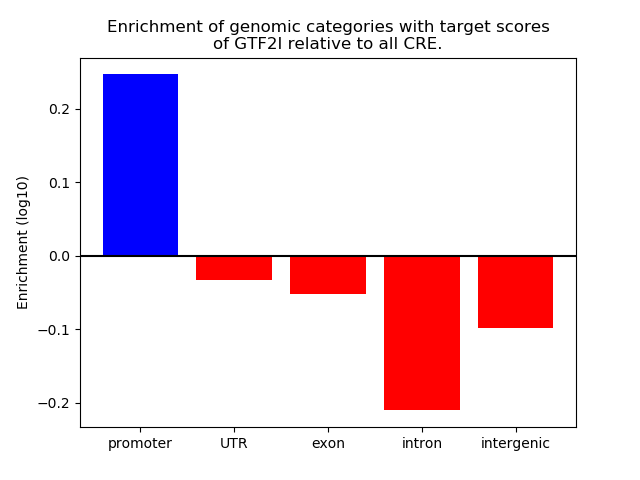
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_57654561_57654712 | 0.29 |
GPR56 |
G protein-coupled receptor 56 |
70 |
0.96 |
| chr12_54367495_54368083 | 0.25 |
HOXC11 |
homeobox C11 |
877 |
0.24 |
| chr19_17905710_17906188 | 0.23 |
B3GNT3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
4 |
0.96 |
| chr11_469322_469620 | 0.23 |
RP13-317D12.3 |
|
5572 |
0.1 |
| chr17_27482435_27482735 | 0.22 |
RP11-321A17.4 |
|
15084 |
0.15 |
| chr18_74798828_74799607 | 0.21 |
MBP |
myelin basic protein |
18000 |
0.27 |
| chr3_184286801_184287672 | 0.21 |
EPHB3 |
EPH receptor B3 |
7664 |
0.2 |
| chr21_32715523_32716836 | 0.20 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
415 |
0.91 |
| chr1_200864498_200864649 | 0.20 |
C1orf106 |
chromosome 1 open reading frame 106 |
624 |
0.71 |
| chr12_54378940_54379308 | 0.18 |
HOXC10 |
homeobox C10 |
154 |
0.46 |
| chr16_29818645_29819422 | 0.17 |
AC009133.15 |
|
20 |
0.51 |
| chr11_116564591_116564742 | 0.17 |
BUD13 |
BUD13 homolog (S. cerevisiae) |
79037 |
0.07 |
| chr19_54984842_54985004 | 0.17 |
CDC42EP5 |
CDC42 effector protein (Rho GTPase binding) 5 |
512 |
0.62 |
| chrX_144509559_144509857 | 0.17 |
SPANXN1 |
SPANX family, member N1 |
181360 |
0.03 |
| chr10_134513988_134514364 | 0.16 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
65152 |
0.12 |
| chr22_46546744_46547896 | 0.16 |
PPARA |
peroxisome proliferator-activated receptor alpha |
467 |
0.75 |
| chr12_85673938_85674715 | 0.16 |
ALX1 |
ALX homeobox 1 |
441 |
0.91 |
| chr2_110872434_110873339 | 0.15 |
MALL |
mal, T-cell differentiation protein-like |
713 |
0.63 |
| chr12_113905433_113905704 | 0.15 |
RP11-82C23.2 |
|
4240 |
0.19 |
| chr1_211688650_211688892 | 0.15 |
RD3 |
retinal degeneration 3 |
22512 |
0.2 |
| chr11_125462781_125463791 | 0.15 |
STT3A |
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
222 |
0.82 |
| chr22_45898330_45898748 | 0.14 |
FBLN1 |
fibulin 1 |
173 |
0.96 |
| chr16_51185618_51185964 | 0.14 |
SALL1 |
spalt-like transcription factor 1 |
605 |
0.76 |
| chr1_156093288_156093439 | 0.14 |
LMNA |
lamin A/C |
2588 |
0.16 |
| chr18_31802690_31803001 | 0.14 |
RP11-379L18.1 |
|
84 |
0.8 |
| chr4_3389662_3389979 | 0.14 |
RGS12 |
regulator of G-protein signaling 12 |
1759 |
0.4 |
| chr1_27924702_27924979 | 0.14 |
AHDC1 |
AT hook, DNA binding motif, containing 1 |
5262 |
0.17 |
| chr16_73081294_73081963 | 0.14 |
ZFHX3 |
zinc finger homeobox 3 |
646 |
0.76 |
| chr1_9895701_9896156 | 0.13 |
CLSTN1 |
calsyntenin 1 |
11344 |
0.16 |
| chr4_116124105_116124256 | 0.13 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
89148 |
0.11 |
| chr4_44451115_44451507 | 0.13 |
KCTD8 |
potassium channel tetramerization domain containing 8 |
487 |
0.76 |
| chr11_126872809_126873319 | 0.13 |
KIRREL3-AS3 |
KIRREL3 antisense RNA 3 |
259 |
0.89 |
| chr12_54391177_54391923 | 0.13 |
HOXC-AS2 |
HOXC cluster antisense RNA 2 |
981 |
0.22 |
| chr17_38500769_38502147 | 0.13 |
RARA |
retinoic acid receptor, alpha |
35 |
0.95 |
| chr11_67078749_67078900 | 0.13 |
SSH3 |
slingshot protein phosphatase 3 |
3696 |
0.13 |
| chr12_85305887_85306488 | 0.13 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
384 |
0.92 |
| chr19_3097315_3097524 | 0.13 |
GNA11 |
guanine nucleotide binding protein (G protein), alpha 11 (Gq class) |
3011 |
0.15 |
| chr19_35606414_35606565 | 0.13 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
243 |
0.84 |
| chr22_25508803_25509083 | 0.13 |
CTA-221G9.11 |
|
284 |
0.93 |
| chr1_214397865_214398571 | 0.13 |
SMYD2 |
SET and MYND domain containing 2 |
56358 |
0.16 |
| chr15_97311897_97312048 | 0.13 |
SPATA8-AS1 |
SPATA8 antisense RNA 1 (head to head) |
12912 |
0.22 |
| chr4_111118316_111119494 | 0.12 |
ELOVL6 |
ELOVL fatty acid elongase 6 |
904 |
0.69 |
| chr16_2517636_2518674 | 0.12 |
RP11-715J22.2 |
|
59 |
0.92 |
| chr9_117050791_117050942 | 0.12 |
ORM1 |
orosomucoid 1 |
34470 |
0.15 |
| chr6_54711000_54712080 | 0.12 |
FAM83B |
family with sequence similarity 83, member B |
29 |
0.98 |
| chr21_47167661_47167812 | 0.12 |
PRED60 |
|
22269 |
0.22 |
| chr10_21462508_21463481 | 0.12 |
NEBL-AS1 |
NEBL antisense RNA 1 |
51 |
0.59 |
| chrX_107681107_107681335 | 0.12 |
COL4A6 |
collagen, type IV, alpha 6 |
335 |
0.88 |
| chr16_52579338_52579756 | 0.12 |
TOX3 |
TOX high mobility group box family member 3 |
1373 |
0.61 |
| chr12_131199797_131200212 | 0.12 |
RIMBP2 |
RIMS binding protein 2 |
822 |
0.73 |
| chr12_6930752_6931082 | 0.12 |
GPR162 |
G protein-coupled receptor 162 |
47 |
0.93 |
| chr19_11494273_11494666 | 0.12 |
EPOR |
erythropoietin receptor |
519 |
0.56 |
| chr20_55205919_55206207 | 0.12 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
125 |
0.96 |
| chr14_103394355_103395516 | 0.12 |
AMN |
amnion associated transmembrane protein |
211 |
0.92 |
| chr16_66443392_66443543 | 0.12 |
CDH5 |
cadherin 5, type 2 (vascular endothelium) |
14109 |
0.16 |
| chr13_114505022_114505231 | 0.12 |
GAS6-AS1 |
GAS6 antisense RNA 1 |
13477 |
0.2 |
| chr16_29910903_29911054 | 0.12 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
110 |
0.86 |
| chr10_5566594_5566745 | 0.12 |
CALML3 |
calmodulin-like 3 |
255 |
0.81 |
| chr3_133748771_133749015 | 0.11 |
SLCO2A1 |
solute carrier organic anion transporter family, member 2A1 |
27 |
0.99 |
| chr3_128207705_128209147 | 0.11 |
RP11-475N22.4 |
|
326 |
0.79 |
| chr11_57365224_57366004 | 0.11 |
SERPING1 |
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
338 |
0.8 |
| chr11_26003807_26004057 | 0.11 |
ANO3 |
anoctamin 3 |
206897 |
0.03 |
| chr20_62573957_62574108 | 0.11 |
ENSG00000207554 |
. |
47 |
0.9 |
| chr10_14215918_14216465 | 0.11 |
RP11-397C18.2 |
|
99908 |
0.08 |
| chr14_38064845_38065186 | 0.11 |
TTC6 |
tetratricopeptide repeat domain 6 |
37 |
0.95 |
| chr6_80656707_80657785 | 0.11 |
ELOVL4 |
ELOVL fatty acid elongase 4 |
51 |
0.98 |
| chr5_32991795_32991946 | 0.11 |
AC026703.1 |
|
202925 |
0.03 |
| chr12_125105824_125106359 | 0.11 |
NCOR2 |
nuclear receptor corepressor 2 |
54081 |
0.17 |
| chr6_5026569_5027044 | 0.11 |
ENSG00000252419 |
. |
19529 |
0.19 |
| chr9_130341109_130341430 | 0.11 |
FAM129B |
family with sequence similarity 129, member B |
1 |
0.98 |
| chr10_77156249_77156959 | 0.11 |
ENSG00000237149 |
. |
4673 |
0.21 |
| chr5_142000582_142000950 | 0.11 |
FGF1 |
fibroblast growth factor 1 (acidic) |
141 |
0.96 |
| chr14_93510273_93510424 | 0.11 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
22355 |
0.18 |
| chr4_144256878_144257439 | 0.11 |
GAB1 |
GRB2-associated binding protein 1 |
757 |
0.67 |
| chr16_73104339_73104801 | 0.11 |
ZFHX3 |
zinc finger homeobox 3 |
10973 |
0.24 |
| chr14_101543718_101544133 | 0.11 |
ENSG00000207959 |
. |
10864 |
0.02 |
| chr2_219155938_219157236 | 0.11 |
TMBIM1 |
transmembrane BAX inhibitor motif containing 1 |
649 |
0.55 |
| chr12_41085967_41086118 | 0.11 |
CNTN1 |
contactin 1 |
202 |
0.97 |
| chr8_42063953_42065062 | 0.11 |
PLAT |
plasminogen activator, tissue |
576 |
0.71 |
| chr12_54784143_54784294 | 0.11 |
ZNF385A |
zinc finger protein 385A |
847 |
0.43 |
| chr19_5261127_5261342 | 0.11 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
24940 |
0.21 |
| chr5_149979916_149980067 | 0.11 |
SYNPO |
synaptopodin |
651 |
0.67 |
| chr4_1035336_1035962 | 0.11 |
FGFRL1 |
fibroblast growth factor receptor-like 1 |
29410 |
0.11 |
| chr15_29966806_29967093 | 0.11 |
RP11-680F8.1 |
|
245 |
0.94 |
| chr19_5880770_5880921 | 0.11 |
FUT5 |
fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
10294 |
0.08 |
| chr10_48355429_48355580 | 0.11 |
ZNF488 |
zinc finger protein 488 |
403 |
0.79 |
| chr11_128812512_128812811 | 0.11 |
TP53AIP1 |
tumor protein p53 regulated apoptosis inducing protein 1 |
161 |
0.95 |
| chr9_109625378_109625771 | 0.11 |
ZNF462 |
zinc finger protein 462 |
127 |
0.97 |
| chr15_43985556_43986365 | 0.10 |
CKMT1A |
creatine kinase, mitochondrial 1A |
109 |
0.93 |
| chr17_71301289_71301523 | 0.10 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
6141 |
0.2 |
| chr9_87283365_87284204 | 0.10 |
NTRK2 |
neurotrophic tyrosine kinase, receptor, type 2 |
318 |
0.95 |
| chr4_4390270_4390555 | 0.10 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
1580 |
0.45 |
| chr22_36232318_36232521 | 0.10 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
3846 |
0.33 |
| chr1_154973351_154974199 | 0.10 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
1352 |
0.2 |
| chr1_13910269_13910760 | 0.10 |
PDPN |
podoplanin |
74 |
0.97 |
| chr19_39280125_39280276 | 0.10 |
LGALS7B |
lectin, galactoside-binding, soluble, 7B |
349 |
0.73 |
| chr16_73033974_73034214 | 0.10 |
AC002044.4 |
|
14281 |
0.2 |
| chr1_156099457_156100025 | 0.10 |
LMNA |
lamin A/C |
3147 |
0.15 |
| chr4_14817384_14817535 | 0.10 |
ENSG00000202449 |
. |
125125 |
0.06 |
| chr12_57998166_57998701 | 0.10 |
DTX3 |
deltex homolog 3 (Drosophila) |
11 |
0.94 |
| chr20_18037263_18037961 | 0.10 |
OVOL2 |
ovo-like zinc finger 2 |
909 |
0.48 |
| chr12_127865548_127865745 | 0.10 |
ENSG00000253089 |
. |
59746 |
0.16 |
| chr8_143693973_143694253 | 0.10 |
ARC |
activity-regulated cytoskeleton-associated protein |
2720 |
0.21 |
| chr5_9782917_9783068 | 0.10 |
ENSG00000222054 |
. |
21973 |
0.24 |
| chr11_395288_395439 | 0.10 |
PKP3 |
plakophilin 3 |
1120 |
0.3 |
| chr1_162666353_162666719 | 0.10 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
64276 |
0.1 |
| chr11_126229473_126229631 | 0.10 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
3683 |
0.19 |
| chr13_114540399_114540680 | 0.10 |
GAS6 |
growth arrest-specific 6 |
1522 |
0.45 |
| chr7_69786510_69786661 | 0.10 |
AUTS2 |
autism susceptibility candidate 2 |
407540 |
0.01 |
| chr2_176961922_176962434 | 0.10 |
HOXD12 |
homeobox D12 |
2280 |
0.13 |
| chr5_132111918_132112304 | 0.10 |
SEPT8 |
septin 8 |
453 |
0.75 |
| chr17_37783095_37783466 | 0.10 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
75 |
0.95 |
| chr11_1221437_1221588 | 0.09 |
MUC5B |
mucin 5B, oligomeric mucus/gel-forming |
22784 |
0.15 |
| chr16_65154751_65155029 | 0.09 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
943 |
0.75 |
| chr4_6472295_6472644 | 0.09 |
PPP2R2C |
protein phosphatase 2, regulatory subunit B, gamma |
1704 |
0.48 |
| chr1_150232422_150232573 | 0.09 |
ENSG00000238526 |
. |
1596 |
0.18 |
| chr14_74100045_74101313 | 0.09 |
RP3-414A15.10 |
|
35 |
0.96 |
| chr15_43885730_43886468 | 0.09 |
CKMT1B |
creatine kinase, mitochondrial 1B |
42 |
0.95 |
| chr7_27197659_27197810 | 0.09 |
HOXA7 |
homeobox A7 |
179 |
0.83 |
| chr1_27357912_27358708 | 0.09 |
FAM46B |
family with sequence similarity 46, member B |
18983 |
0.16 |
| chr2_91846908_91847477 | 0.09 |
AC027612.6 |
|
783 |
0.68 |
| chr5_172721162_172721383 | 0.09 |
ENSG00000252169 |
. |
1650 |
0.36 |
| chr12_112818867_112820166 | 0.09 |
HECTD4 |
HECT domain containing E3 ubiquitin protein ligase 4 |
380 |
0.87 |
| chr7_150822785_150823007 | 0.09 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
7600 |
0.11 |
| chr2_121199524_121199751 | 0.09 |
LINC01101 |
long intergenic non-protein coding RNA 1101 |
24060 |
0.24 |
| chr1_3615954_3616105 | 0.09 |
TP73 |
tumor protein p73 |
1407 |
0.29 |
| chr12_52919023_52919174 | 0.09 |
AC055736.1 |
Uncharacterized protein |
3817 |
0.13 |
| chr17_41476988_41477313 | 0.09 |
ARL4D |
ADP-ribosylation factor-like 4D |
823 |
0.46 |
| chr6_156488488_156489372 | 0.09 |
ENSG00000212295 |
. |
210954 |
0.02 |
| chr2_242499136_242499442 | 0.09 |
BOK-AS1 |
BOK antisense RNA 1 |
897 |
0.38 |
| chr17_46674549_46675551 | 0.09 |
HOXB-AS3 |
HOXB cluster antisense RNA 3 |
1249 |
0.2 |
| chr15_96897591_96898313 | 0.09 |
AC087477.1 |
Uncharacterized protein |
6535 |
0.18 |
| chr22_43115644_43116713 | 0.09 |
A4GALT |
alpha 1,4-galactosyltransferase |
24539 |
0.15 |
| chr8_1778332_1778483 | 0.09 |
ARHGEF10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
4130 |
0.24 |
| chr11_57089871_57090022 | 0.09 |
TNKS1BP1 |
tankyrase 1 binding protein 1, 182kDa |
137 |
0.92 |
| chr14_85999097_85999705 | 0.09 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
2829 |
0.32 |
| chr1_56111785_56112048 | 0.09 |
ENSG00000272051 |
. |
161271 |
0.04 |
| chr2_172959441_172959633 | 0.09 |
DLX2 |
distal-less homeobox 2 |
8091 |
0.21 |
| chr2_11123558_11123709 | 0.09 |
KCNF1 |
potassium voltage-gated channel, subfamily F, member 1 |
71570 |
0.1 |
| chr9_1050369_1050635 | 0.09 |
DMRT2 |
doublesex and mab-3 related transcription factor 2 |
118 |
0.98 |
| chr6_35360979_35361130 | 0.09 |
PPARD |
peroxisome proliferator-activated receptor delta |
50652 |
0.11 |
| chr12_50450066_50450217 | 0.09 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
1190 |
0.36 |
| chr7_31781380_31781531 | 0.09 |
ENSG00000223070 |
. |
52407 |
0.15 |
| chr17_77030183_77030334 | 0.09 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
9 |
0.97 |
| chr14_100004080_100004231 | 0.09 |
CCDC85C |
coiled-coil domain containing 85C |
1730 |
0.33 |
| chr15_70702879_70703030 | 0.09 |
ENSG00000200216 |
. |
217379 |
0.02 |
| chr19_40785123_40785301 | 0.09 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
334 |
0.77 |
| chr15_71036630_71036796 | 0.09 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
19056 |
0.21 |
| chr15_37387605_37387763 | 0.09 |
MEIS2 |
Meis homeobox 2 |
462 |
0.84 |
| chr9_124517266_124517417 | 0.09 |
DAB2IP |
DAB2 interacting protein |
12277 |
0.25 |
| chr12_52627039_52627830 | 0.09 |
KRT7 |
keratin 7 |
536 |
0.63 |
| chr1_41898610_41898871 | 0.09 |
ENSG00000252563 |
. |
33959 |
0.16 |
| chr4_20703224_20703375 | 0.09 |
PACRGL |
PARK2 co-regulated-like |
1195 |
0.6 |
| chr2_99954548_99954935 | 0.09 |
EIF5B |
eukaryotic translation initiation factor 5B |
925 |
0.48 |
| chr11_1220573_1220724 | 0.09 |
MUC5B |
mucin 5B, oligomeric mucus/gel-forming |
23648 |
0.15 |
| chr1_161167898_161168112 | 0.09 |
ADAMTS4 |
ADAM metallopeptidase with thrombospondin type 1 motif, 4 |
838 |
0.28 |
| chr19_10736216_10736429 | 0.09 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
121 |
0.93 |
| chrX_133147131_133147282 | 0.09 |
GPC3 |
glypican 3 |
27284 |
0.21 |
| chr2_73384123_73384274 | 0.09 |
RAB11FIP5 |
RAB11 family interacting protein 5 (class I) |
44052 |
0.1 |
| chr2_232538307_232538458 | 0.09 |
ENSG00000239202 |
. |
27398 |
0.13 |
| chr4_23975239_23975390 | 0.09 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
83614 |
0.11 |
| chr7_95403720_95403871 | 0.09 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
1879 |
0.48 |
| chr8_75232625_75233066 | 0.09 |
JPH1 |
junctophilin 1 |
718 |
0.68 |
| chr2_106886104_106886958 | 0.09 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
75736 |
0.1 |
| chr9_140199344_140199495 | 0.09 |
NRARP |
NOTCH-regulated ankyrin repeat protein |
2716 |
0.14 |
| chr18_77929070_77929234 | 0.09 |
AC139100.3 |
|
12411 |
0.18 |
| chr3_159481083_159481637 | 0.09 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
142 |
0.96 |
| chr2_56412283_56412467 | 0.09 |
AC007743.1 |
|
327 |
0.83 |
| chr18_10560102_10560331 | 0.09 |
NAPG |
N-ethylmaleimide-sensitive factor attachment protein, gamma |
33807 |
0.19 |
| chr19_39263775_39263926 | 0.09 |
LGALS7 |
lectin, galactoside-binding, soluble, 7 |
222 |
0.85 |
| chr1_44399484_44399885 | 0.09 |
ARTN |
artemin |
166 |
0.92 |
| chr14_86000288_86000530 | 0.09 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
3837 |
0.29 |
| chr1_16472071_16472222 | 0.09 |
RP11-276H7.2 |
|
9560 |
0.12 |
| chr22_31644271_31644422 | 0.09 |
LIMK2 |
LIM domain kinase 2 |
2 |
0.96 |
| chr3_138663757_138663928 | 0.09 |
FOXL2 |
forkhead box L2 |
2140 |
0.26 |
| chr18_8983307_8983458 | 0.09 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
119246 |
0.05 |
| chr13_27334023_27334664 | 0.09 |
GPR12 |
G protein-coupled receptor 12 |
84 |
0.98 |
| chr7_27153769_27154326 | 0.09 |
HOXA-AS2 |
HOXA cluster antisense RNA 2 |
492 |
0.41 |
| chr3_49170263_49170561 | 0.09 |
LAMB2 |
laminin, beta 2 (laminin S) |
41 |
0.95 |
| chr1_45083000_45083241 | 0.09 |
RNF220 |
ring finger protein 220 |
8878 |
0.17 |
| chr17_78165764_78166017 | 0.09 |
CARD14 |
caspase recruitment domain family, member 14 |
7916 |
0.14 |
| chr22_46980499_46980650 | 0.09 |
GRAMD4 |
GRAM domain containing 4 |
7548 |
0.22 |
| chr22_29704652_29704931 | 0.09 |
GAS2L1 |
growth arrest-specific 2 like 1 |
1743 |
0.22 |
| chr18_24083489_24083640 | 0.09 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
43399 |
0.17 |
| chr9_133539320_133539471 | 0.09 |
PRDM12 |
PR domain containing 12 |
586 |
0.77 |
| chr10_13889361_13889512 | 0.08 |
FRMD4A |
FERM domain containing 4A |
11476 |
0.24 |
| chr16_84676084_84676235 | 0.08 |
KLHL36 |
kelch-like family member 36 |
5972 |
0.19 |
| chr22_47135360_47135511 | 0.08 |
CERK |
ceramide kinase |
1277 |
0.45 |
| chr19_4813287_4813441 | 0.08 |
AC005523.2 |
|
17793 |
0.1 |
| chr1_228337001_228337152 | 0.08 |
GJC2 |
gap junction protein, gamma 2, 47kDa |
477 |
0.67 |
| chr17_56494195_56494346 | 0.08 |
RNF43 |
ring finger protein 43 |
176 |
0.93 |
| chr1_27190573_27190827 | 0.08 |
SFN |
stratifin |
1067 |
0.35 |
| chr20_32899455_32899940 | 0.08 |
AHCY |
adenosylhomocysteinase |
89 |
0.96 |
| chr2_182521265_182521506 | 0.08 |
CERKL |
ceramide kinase-like |
348 |
0.89 |
| chr8_104511456_104511779 | 0.08 |
RP11-1C8.4 |
|
169 |
0.93 |
| chr5_73502284_73502435 | 0.08 |
ENSG00000222551 |
. |
65231 |
0.14 |
| chr1_185703154_185703536 | 0.08 |
HMCN1 |
hemicentin 1 |
338 |
0.93 |
| chr16_30022591_30022974 | 0.08 |
DOC2A |
double C2-like domains, alpha |
40 |
0.94 |
| chr10_123357016_123357770 | 0.08 |
FGFR2 |
fibroblast growth factor receptor 2 |
205 |
0.97 |
| chrX_9754895_9755291 | 0.08 |
SHROOM2 |
shroom family member 2 |
597 |
0.54 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 0.2 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.1 | 0.2 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.2 | GO:0036303 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.0 | 0.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.0 | GO:0009111 | vitamin catabolic process(GO:0009111) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003337) metanephric renal vesicle morphogenesis(GO:0072283) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.2 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.0 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.0 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.0 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.2 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.0 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.0 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.0 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0003170 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.0 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:0072109 | glomerular mesangium development(GO:0072109) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:0060463 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.2 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.2 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.0 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.0 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.0 | GO:0072176 | nephric duct development(GO:0072176) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.0 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.0 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.0 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |