Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
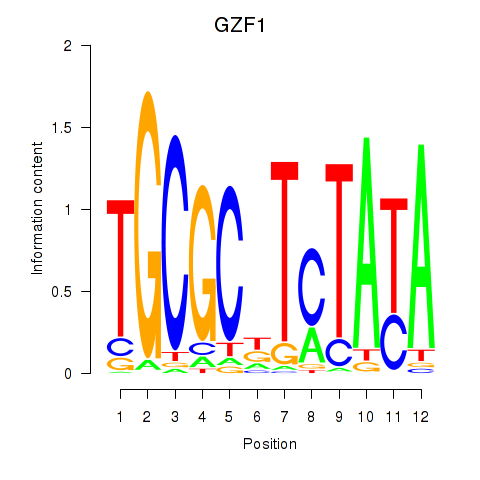
Results for GZF1
Z-value: 0.78
Transcription factors associated with GZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GZF1
|
ENSG00000125812.11 | GDNF inducible zinc finger protein 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr20_23342076_23342460 | GZF1 | 519 | 0.643610 | 0.69 | 3.8e-02 | Click! |
| chr20_23346779_23346930 | GZF1 | 1833 | 0.210527 | -0.67 | 5.0e-02 | Click! |
| chr20_23343011_23343833 | GZF1 | 465 | 0.690043 | 0.32 | 4.0e-01 | Click! |
Activity of the GZF1 motif across conditions
Conditions sorted by the z-value of the GZF1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_118652704_118652966 | 0.44 |
ENO4 |
enolase family member 4 |
34290 |
0.17 |
| chr16_1257169_1257320 | 0.42 |
RP11-616M22.3 |
|
120 |
0.92 |
| chr16_375168_375449 | 0.41 |
AXIN1 |
axin 1 |
27141 |
0.09 |
| chr2_29537347_29537508 | 0.36 |
ALK |
anaplastic lymphoma receptor tyrosine kinase |
91167 |
0.08 |
| chr12_116985994_116986414 | 0.30 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
10982 |
0.28 |
| chr20_42086642_42086854 | 0.30 |
SRSF6 |
serine/arginine-rich splicing factor 6 |
180 |
0.93 |
| chr10_104005591_104006435 | 0.29 |
GBF1 |
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
724 |
0.58 |
| chr10_14698721_14698872 | 0.29 |
RP11-7C6.1 |
|
4055 |
0.23 |
| chr7_64712185_64712407 | 0.29 |
ENSG00000265175 |
. |
16013 |
0.22 |
| chr12_32115176_32115327 | 0.27 |
KIAA1551 |
KIAA1551 |
172 |
0.96 |
| chr6_157342303_157342500 | 0.27 |
RP1-137K2.2 |
|
98439 |
0.08 |
| chr8_142427875_142428034 | 0.27 |
PTP4A3 |
protein tyrosine phosphatase type IVA, member 3 |
4053 |
0.16 |
| chr8_21916945_21917106 | 0.26 |
DMTN |
dematin actin binding protein |
291 |
0.84 |
| chr11_96656080_96656231 | 0.25 |
ENSG00000200411 |
. |
448299 |
0.01 |
| chr5_126323849_126324159 | 0.25 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
42496 |
0.18 |
| chr2_27603785_27604012 | 0.25 |
ZNF513 |
zinc finger protein 513 |
241 |
0.83 |
| chr12_53846681_53847472 | 0.25 |
PCBP2 |
poly(rC) binding protein 2 |
448 |
0.72 |
| chr5_132158669_132158820 | 0.24 |
SHROOM1 |
shroom family member 1 |
3088 |
0.15 |
| chr14_75894369_75894594 | 0.24 |
RP11-293M10.6 |
|
37 |
0.52 |
| chrX_75392544_75392813 | 0.24 |
PBDC1 |
polysaccharide biosynthesis domain containing 1 |
93 |
0.98 |
| chr9_133556690_133556909 | 0.23 |
EXOSC2 |
exosome component 2 |
12309 |
0.18 |
| chr12_11699256_11699407 | 0.22 |
ENSG00000251747 |
. |
82 |
0.97 |
| chr14_21979649_21979818 | 0.22 |
METTL3 |
methyltransferase like 3 |
216 |
0.9 |
| chr14_52739518_52739669 | 0.22 |
PTGDR |
prostaglandin D2 receptor (DP) |
5063 |
0.28 |
| chr1_153643822_153644080 | 0.22 |
ILF2 |
interleukin enhancer binding factor 2 |
427 |
0.65 |
| chr14_106495447_106495598 | 0.22 |
IGHVIII-5-1 |
immunoglobulin heavy variable (III)-5-1 (pseudogene) |
474 |
0.43 |
| chr20_34077433_34077600 | 0.22 |
RP3-477O4.14 |
|
1227 |
0.32 |
| chr19_49148382_49148554 | 0.21 |
CA11 |
carbonic anhydrase XI |
1101 |
0.28 |
| chr9_139743848_139744034 | 0.21 |
PHPT1 |
phosphohistidine phosphatase 1 |
395 |
0.63 |
| chrX_129535378_129535825 | 0.20 |
RBMX2 |
RNA binding motif protein, X-linked 2 |
342 |
0.89 |
| chr1_27683876_27684079 | 0.20 |
MAP3K6 |
mitogen-activated protein kinase kinase kinase 6 |
1015 |
0.4 |
| chr19_3557088_3557331 | 0.20 |
MFSD12 |
major facilitator superfamily domain containing 12 |
192 |
0.87 |
| chr7_45019061_45019212 | 0.20 |
MYO1G |
myosin IG |
439 |
0.76 |
| chr21_15916676_15916915 | 0.20 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
1867 |
0.42 |
| chr3_45017798_45018180 | 0.20 |
EXOSC7 |
exosome component 7 |
267 |
0.53 |
| chr1_197170699_197171135 | 0.19 |
CRB1 |
crumbs homolog 1 (Drosophila) |
325 |
0.84 |
| chr6_106545216_106545824 | 0.19 |
PRDM1 |
PR domain containing 1, with ZNF domain |
961 |
0.58 |
| chr8_117885462_117886204 | 0.19 |
RAD21 |
RAD21 homolog (S. pombe) |
766 |
0.36 |
| chr12_120662827_120663239 | 0.19 |
PXN |
paxillin |
480 |
0.71 |
| chr2_187351379_187351888 | 0.19 |
ZC3H15 |
zinc finger CCCH-type containing 15 |
653 |
0.72 |
| chr3_156393455_156393974 | 0.19 |
TIPARP |
TCDD-inducible poly(ADP-ribose) polymerase |
356 |
0.85 |
| chr12_3162069_3162220 | 0.19 |
TSPAN9 |
tetraspanin 9 |
24377 |
0.17 |
| chr1_199878058_199878346 | 0.18 |
ENSG00000200139 |
. |
20915 |
0.24 |
| chr1_38260135_38260459 | 0.18 |
MANEAL |
mannosidase, endo-alpha-like |
142 |
0.91 |
| chr6_122792252_122792563 | 0.18 |
SERINC1 |
serine incorporator 1 |
560 |
0.52 |
| chr14_50319523_50319960 | 0.18 |
NEMF |
nuclear export mediator factor |
180 |
0.85 |
| chr8_6566608_6567048 | 0.18 |
AGPAT5 |
1-acylglycerol-3-phosphate O-acyltransferase 5 |
614 |
0.61 |
| chr5_67580975_67581126 | 0.18 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
3146 |
0.37 |
| chr3_52718835_52719171 | 0.18 |
PBRM1 |
polybromo 1 |
591 |
0.42 |
| chrX_106956955_106957175 | 0.18 |
TSC22D3 |
TSC22 domain family, member 3 |
2566 |
0.34 |
| chr6_89789489_89789998 | 0.18 |
RP11-63L7.5 |
|
643 |
0.43 |
| chr12_127630819_127631001 | 0.18 |
ENSG00000239776 |
. |
19762 |
0.3 |
| chr2_28836714_28836865 | 0.18 |
PLB1 |
phospholipase B1 |
12003 |
0.21 |
| chr14_102227472_102227811 | 0.17 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
494 |
0.78 |
| chr7_36305572_36305723 | 0.17 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
31106 |
0.15 |
| chr8_21769341_21769492 | 0.17 |
DOK2 |
docking protein 2, 56kDa |
1758 |
0.36 |
| chr11_36423391_36423542 | 0.17 |
PRR5L |
proline rich 5 like |
851 |
0.61 |
| chr22_30662105_30662275 | 0.17 |
OSM |
oncostatin M |
383 |
0.75 |
| chr12_104458290_104458598 | 0.16 |
HCFC2 |
host cell factor C2 |
148 |
0.82 |
| chr2_134877443_134877671 | 0.16 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
196 |
0.96 |
| chr8_29953492_29954050 | 0.16 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
606 |
0.68 |
| chr5_473388_473548 | 0.16 |
ENSG00000225138 |
. |
117 |
0.68 |
| chr7_105362356_105362507 | 0.16 |
ATXN7L1 |
ataxin 7-like 1 |
30347 |
0.23 |
| chr6_34112105_34112270 | 0.16 |
GRM4 |
glutamate receptor, metabotropic 4 |
1670 |
0.44 |
| chr21_38073571_38073742 | 0.16 |
AP000697.6 |
|
208 |
0.93 |
| chr16_89557343_89557542 | 0.16 |
ANKRD11 |
ankyrin repeat domain 11 |
473 |
0.69 |
| chr3_158390470_158390795 | 0.16 |
LXN |
latexin |
150 |
0.95 |
| chr7_156803754_156804127 | 0.15 |
MNX1-AS1 |
MNX1 antisense RNA 1 (head to head) |
441 |
0.56 |
| chr1_92946934_92947085 | 0.15 |
GFI1 |
growth factor independent 1 transcription repressor |
2502 |
0.37 |
| chr22_31619723_31619874 | 0.15 |
ENSG00000199695 |
. |
959 |
0.4 |
| chrX_102631036_102631748 | 0.15 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
57 |
0.97 |
| chr10_103879598_103879834 | 0.15 |
LDB1 |
LIM domain binding 1 |
494 |
0.73 |
| chr3_81472677_81472828 | 0.15 |
ENSG00000222389 |
. |
85875 |
0.11 |
| chr16_67876459_67876770 | 0.15 |
THAP11 |
THAP domain containing 11 |
401 |
0.44 |
| chr12_122326739_122327128 | 0.15 |
PSMD9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
241 |
0.9 |
| chr9_135117163_135117384 | 0.15 |
ENSG00000221105 |
. |
27011 |
0.2 |
| chr5_170814799_170815118 | 0.15 |
NPM1 |
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
73 |
0.95 |
| chr17_73773061_73773428 | 0.15 |
H3F3B |
H3 histone, family 3B (H3.3B) |
2123 |
0.16 |
| chr19_58987134_58987390 | 0.14 |
CTD-2619J13.23 |
|
132 |
0.64 |
| chr12_125473806_125474002 | 0.14 |
DHX37 |
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
237 |
0.93 |
| chr20_61550921_61551072 | 0.14 |
DIDO1 |
death inducer-obliterator 1 |
5238 |
0.16 |
| chr6_167411127_167411495 | 0.14 |
ENSG00000265828 |
. |
89 |
0.93 |
| chr16_68452108_68452259 | 0.14 |
SMPD3 |
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
30212 |
0.12 |
| chr1_239551018_239551169 | 0.14 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
241280 |
0.02 |
| chr3_135969234_135969511 | 0.14 |
PCCB |
propionyl CoA carboxylase, beta polypeptide |
155 |
0.97 |
| chr15_31765016_31765167 | 0.14 |
KLF13 |
Kruppel-like factor 13 |
106734 |
0.07 |
| chr4_82135724_82136079 | 0.14 |
PRKG2 |
protein kinase, cGMP-dependent, type II |
287 |
0.94 |
| chr13_53192037_53192369 | 0.14 |
HNRNPA1L2 |
heterogeneous nuclear ribonucleoprotein A1-like 2 |
598 |
0.79 |
| chr8_135844464_135844657 | 0.14 |
ENSG00000199153 |
. |
27372 |
0.21 |
| chr21_36262594_36262745 | 0.14 |
RUNX1 |
runt-related transcription factor 1 |
582 |
0.85 |
| chr20_62612476_62612847 | 0.14 |
PRPF6 |
pre-mRNA processing factor 6 |
173 |
0.87 |
| chr9_74378842_74379095 | 0.14 |
TMEM2 |
transmembrane protein 2 |
4334 |
0.34 |
| chr6_57181574_57181908 | 0.13 |
PRIM2 |
primase, DNA, polypeptide 2 (58kDa) |
681 |
0.78 |
| chr6_42418365_42418744 | 0.13 |
TRERF1 |
transcriptional regulating factor 1 |
445 |
0.85 |
| chr6_7108626_7109201 | 0.13 |
RREB1 |
ras responsive element binding protein 1 |
649 |
0.77 |
| chr9_95877171_95877322 | 0.13 |
C9orf89 |
chromosome 9 open reading frame 89 |
18746 |
0.14 |
| chr6_30524774_30525039 | 0.13 |
GNL1 |
guanine nucleotide binding protein-like 1 |
45 |
0.63 |
| chr5_133449213_133449364 | 0.13 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
1114 |
0.56 |
| chr14_36788990_36789756 | 0.13 |
MBIP |
MAP3K12 binding inhibitory protein 1 |
297 |
0.92 |
| chr11_2502582_2502733 | 0.13 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
19540 |
0.16 |
| chr12_90378520_90378671 | 0.13 |
ENSG00000252823 |
. |
230759 |
0.02 |
| chr10_106083791_106084156 | 0.13 |
ITPRIP |
inositol 1,4,5-trisphosphate receptor interacting protein |
4709 |
0.15 |
| chr18_60988479_60988630 | 0.13 |
BCL2 |
B-cell CLL/lymphoma 2 |
1193 |
0.44 |
| chr13_73355012_73355163 | 0.13 |
DIS3 |
DIS3 mitotic control homolog (S. cerevisiae) |
976 |
0.39 |
| chr9_80625845_80626079 | 0.13 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
19558 |
0.28 |
| chr21_42258267_42258418 | 0.13 |
DSCAM |
Down syndrome cell adhesion molecule |
39277 |
0.21 |
| chrX_153618503_153618757 | 0.13 |
RPL10 |
ribosomal protein L10 |
6865 |
0.08 |
| chrX_75365570_75365805 | 0.12 |
PBDC1 |
polysaccharide biosynthesis domain containing 1 |
27084 |
0.25 |
| chr6_109800480_109800632 | 0.12 |
ZBTB24 |
zinc finger and BTB domain containing 24 |
3884 |
0.14 |
| chr1_27951925_27952076 | 0.12 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
646 |
0.63 |
| chr12_11803481_11804350 | 0.12 |
ETV6 |
ets variant 6 |
1127 |
0.62 |
| chr2_203775980_203776689 | 0.12 |
CARF |
calcium responsive transcription factor |
603 |
0.45 |
| chr2_220042618_220042887 | 0.12 |
CNPPD1 |
cyclin Pas1/PHO80 domain containing 1 |
76 |
0.53 |
| chr5_175962769_175963088 | 0.12 |
RNF44 |
ring finger protein 44 |
1493 |
0.28 |
| chr3_52007652_52007993 | 0.12 |
ABHD14A |
abhydrolase domain containing 14A |
122 |
0.44 |
| chr3_185654118_185654306 | 0.12 |
TRA2B |
transformer 2 beta homolog (Drosophila) |
1589 |
0.4 |
| chr4_155220038_155220189 | 0.12 |
ENSG00000264137 |
. |
80052 |
0.1 |
| chr1_206135676_206135961 | 0.12 |
FAM72A |
family with sequence similarity 72, member A |
1419 |
0.38 |
| chr1_26758171_26758572 | 0.12 |
DHDDS |
dehydrodolichyl diphosphate synthase |
436 |
0.76 |
| chr8_146175786_146175956 | 0.12 |
ZNF16 |
zinc finger protein 16 |
110 |
0.96 |
| chr5_149521280_149521431 | 0.12 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
4355 |
0.17 |
| chr10_104005118_104005269 | 0.12 |
GBF1 |
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
96 |
0.95 |
| chr17_56407127_56407278 | 0.12 |
BZRAP1-AS1 |
BZRAP1 antisense RNA 1 |
236 |
0.8 |
| chrX_153570073_153570387 | 0.12 |
XX-FW83128A1.2 |
|
6843 |
0.1 |
| chr2_27008209_27008614 | 0.12 |
CENPA |
centromere protein A |
454 |
0.81 |
| chr1_1004234_1004415 | 0.12 |
RNF223 |
ring finger protein 223 |
5363 |
0.1 |
| chr6_24774737_24775060 | 0.12 |
GMNN |
geminin, DNA replication inhibitor |
261 |
0.9 |
| chr12_9903163_9903484 | 0.12 |
CD69 |
CD69 molecule |
10174 |
0.15 |
| chr17_62222536_62222687 | 0.12 |
ENSG00000199753 |
. |
832 |
0.44 |
| chr9_108319921_108320392 | 0.12 |
RP11-235C23.5 |
|
154 |
0.64 |
| chr1_202310113_202310440 | 0.12 |
UBE2T |
ubiquitin-conjugating enzyme E2T (putative) |
832 |
0.62 |
| chr2_106777289_106777491 | 0.12 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
309 |
0.92 |
| chr3_120068350_120069016 | 0.12 |
RP11-174O3.3 |
|
326 |
0.63 |
| chr14_70041081_70041336 | 0.12 |
CCDC177 |
Homo sapiens coiled-coil domain containing 177 (CCDC177), mRNA. |
869 |
0.67 |
| chr20_10903511_10903679 | 0.12 |
RP11-103J8.1 |
|
20663 |
0.27 |
| chr11_36531965_36532300 | 0.12 |
TRAF6 |
TNF receptor-associated factor 6, E3 ubiquitin protein ligase |
310 |
0.91 |
| chr19_4633711_4633862 | 0.11 |
TNFAIP8L1 |
tumor necrosis factor, alpha-induced protein 8-like 1 |
5744 |
0.13 |
| chr11_118489320_118489642 | 0.11 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
11123 |
0.11 |
| chr22_32806330_32806481 | 0.11 |
RTCB |
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
1819 |
0.29 |
| chr3_48693068_48693287 | 0.11 |
CELSR3 |
cadherin, EGF LAG seven-pass G-type receptor 3 |
7171 |
0.09 |
| chr13_46627012_46627449 | 0.11 |
CPB2-AS1 |
CPB2 antisense RNA 1 |
193 |
0.67 |
| chr12_8724646_8724797 | 0.11 |
ENSG00000238943 |
. |
20351 |
0.12 |
| chr1_7765903_7766129 | 0.11 |
CAMTA1 |
calmodulin binding transcription activator 1 |
30452 |
0.17 |
| chr22_43355548_43355783 | 0.11 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
219 |
0.95 |
| chr12_102513335_102513786 | 0.11 |
NUP37 |
nucleoporin 37kDa |
338 |
0.53 |
| chr1_149813796_149814193 | 0.11 |
HIST2H2AA3 |
histone cluster 2, H2aa3 |
484 |
0.49 |
| chr19_2237563_2237785 | 0.11 |
PLEKHJ1 |
pleckstrin homology domain containing, family J member 1 |
29 |
0.59 |
| chr7_72936777_72937003 | 0.11 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
282 |
0.88 |
| chr20_5093003_5093662 | 0.11 |
TMEM230 |
transmembrane protein 230 |
67 |
0.94 |
| chr16_11763075_11763345 | 0.11 |
SNN |
stannin |
940 |
0.56 |
| chr5_162934649_162934884 | 0.11 |
MAT2B |
methionine adenosyltransferase II, beta |
2146 |
0.28 |
| chr5_55287822_55288099 | 0.11 |
IL6ST |
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
2832 |
0.2 |
| chr2_30448704_30448864 | 0.11 |
LBH |
limb bud and heart development |
5613 |
0.24 |
| chr11_118868535_118868695 | 0.11 |
CCDC84 |
coiled-coil domain containing 84 |
237 |
0.79 |
| chr17_74137986_74138137 | 0.11 |
RNF157-AS1 |
RNF157 antisense RNA 1 |
464 |
0.54 |
| chr6_27841415_27841672 | 0.11 |
HIST1H4L |
histone cluster 1, H4l |
254 |
0.76 |
| chr14_50087068_50087296 | 0.11 |
RPL36AL |
ribosomal protein L36a-like |
221 |
0.56 |
| chr10_103576027_103576601 | 0.11 |
MGEA5 |
meningioma expressed antigen 5 (hyaluronidase) |
1309 |
0.34 |
| chr8_144373646_144373987 | 0.11 |
ZNF696 |
zinc finger protein 696 |
224 |
0.82 |
| chr10_22541853_22542149 | 0.11 |
EBLN1 |
endogenous Bornavirus-like nucleoprotein 1 |
43051 |
0.13 |
| chrX_154445779_154445930 | 0.11 |
VBP1 |
von Hippel-Lindau binding protein 1 |
1211 |
0.48 |
| chr21_34864166_34864389 | 0.11 |
DNAJC28 |
DnaJ (Hsp40) homolog, subfamily C, member 28 |
250 |
0.87 |
| chr16_46723241_46723448 | 0.11 |
ORC6 |
origin recognition complex, subunit 6 |
214 |
0.52 |
| chr7_65762308_65762459 | 0.11 |
ENSG00000252126 |
. |
46908 |
0.14 |
| chr16_22217946_22218447 | 0.11 |
EEF2K |
eukaryotic elongation factor-2 kinase |
593 |
0.74 |
| chr1_144593335_144593591 | 0.11 |
RP11-640M9.2 |
|
21668 |
0.2 |
| chr6_88412249_88412889 | 0.11 |
AKIRIN2 |
akirin 2 |
642 |
0.73 |
| chr6_11094046_11094248 | 0.10 |
SMIM13 |
small integral membrane protein 13 |
119 |
0.96 |
| chr5_168729442_168729593 | 0.10 |
SLIT3 |
slit homolog 3 (Drosophila) |
1384 |
0.54 |
| chr17_18087734_18087959 | 0.10 |
RP11-258F1.1 |
|
135 |
0.69 |
| chrX_24072030_24072308 | 0.10 |
EIF2S3 |
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
664 |
0.73 |
| chr11_111957062_111957410 | 0.10 |
SDHD |
succinate dehydrogenase complex, subunit D, integral membrane protein |
261 |
0.49 |
| chr3_176915816_176916502 | 0.10 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
898 |
0.7 |
| chr9_131133752_131134483 | 0.10 |
URM1 |
ubiquitin related modifier 1 |
467 |
0.68 |
| chr2_43449640_43450246 | 0.10 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
3805 |
0.27 |
| chr3_171176304_171176455 | 0.10 |
TNIK |
TRAF2 and NCK interacting kinase |
1473 |
0.45 |
| chrX_75368988_75369430 | 0.10 |
PBDC1 |
polysaccharide biosynthesis domain containing 1 |
23562 |
0.26 |
| chr1_192842951_192843102 | 0.10 |
ENSG00000223075 |
. |
2089 |
0.41 |
| chr6_159072646_159072797 | 0.10 |
SYTL3 |
synaptotagmin-like 3 |
1675 |
0.35 |
| chr6_64345134_64345489 | 0.10 |
PHF3 |
PHD finger protein 3 |
414 |
0.89 |
| chr19_53104607_53104836 | 0.10 |
CTD-3099C6.11 |
|
2581 |
0.16 |
| chr12_14926776_14927159 | 0.10 |
H2AFJ |
H2A histone family, member J |
303 |
0.82 |
| chrX_9866552_9866703 | 0.10 |
SHROOM2 |
shroom family member 2 |
13785 |
0.2 |
| chr22_32057584_32058186 | 0.10 |
PISD |
phosphatidylserine decarboxylase |
281 |
0.92 |
| chr13_52767860_52768554 | 0.10 |
MRPS31P5 |
mitochondrial ribosomal protein S31 pseudogene 5 |
306 |
0.91 |
| chr5_35857770_35858187 | 0.10 |
IL7R |
interleukin 7 receptor |
984 |
0.58 |
| chr11_122050649_122050906 | 0.10 |
ENSG00000207994 |
. |
27761 |
0.16 |
| chr11_62529664_62529976 | 0.10 |
POLR2G |
polymerase (RNA) II (DNA directed) polypeptide G |
783 |
0.32 |
| chr3_16351051_16351202 | 0.10 |
RP11-415F23.2 |
|
4820 |
0.19 |
| chr1_241840893_241841044 | 0.10 |
WDR64 |
WD repeat domain 64 |
5914 |
0.23 |
| chrX_124911556_124911707 | 0.10 |
ENSG00000222518 |
. |
114732 |
0.07 |
| chr1_42181439_42181590 | 0.10 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
14842 |
0.25 |
| chr8_108509260_108510202 | 0.10 |
ANGPT1 |
angiopoietin 1 |
507 |
0.89 |
| chr22_51021623_51022044 | 0.10 |
CHKB-AS1 |
CHKB antisense RNA 1 (head to head) |
378 |
0.43 |
| chr6_501893_502410 | 0.10 |
RP1-20B11.2 |
|
22020 |
0.26 |
| chrX_41280637_41281001 | 0.10 |
NYX |
nyctalopin |
25868 |
0.18 |
| chr6_47381977_47382964 | 0.10 |
ENSG00000266330 |
. |
48182 |
0.14 |
| chr9_137217336_137217895 | 0.10 |
RXRA |
retinoid X receptor, alpha |
811 |
0.72 |
| chr9_136999905_137000059 | 0.10 |
WDR5 |
WD repeat domain 5 |
505 |
0.59 |
| chr4_123072364_123072515 | 0.10 |
KIAA1109 |
KIAA1109 |
1049 |
0.65 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.2 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.2 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.0 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0097205 | regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0046885 | regulation of hormone biosynthetic process(GO:0046885) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.2 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0042511 | positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.0 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.0 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |