Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
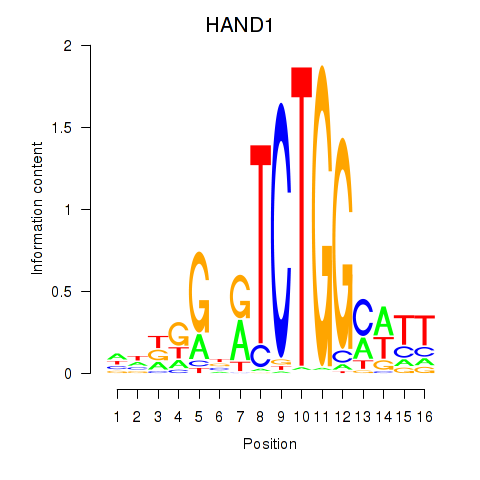
Results for HAND1
Z-value: 1.14
Transcription factors associated with HAND1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HAND1
|
ENSG00000113196.2 | heart and neural crest derivatives expressed 1 |
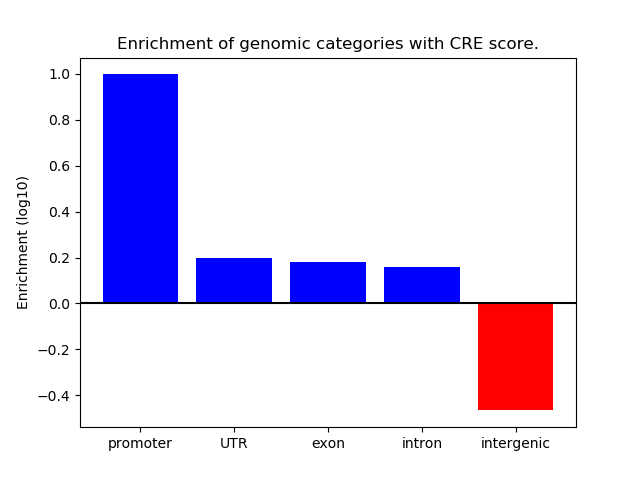
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_153903378_153903529 | HAND1 | 45629 | 0.128195 | -0.82 | 6.8e-03 | Click! |
| chr5_153900789_153900940 | HAND1 | 43040 | 0.133580 | -0.73 | 2.5e-02 | Click! |
| chr5_153856732_153856883 | HAND1 | 1017 | 0.530656 | 0.65 | 5.6e-02 | Click! |
| chr5_153873035_153873186 | HAND1 | 15286 | 0.183386 | 0.56 | 1.2e-01 | Click! |
| chr5_153857033_153857184 | HAND1 | 716 | 0.669615 | 0.53 | 1.5e-01 | Click! |
Activity of the HAND1 motif across conditions
Conditions sorted by the z-value of the HAND1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
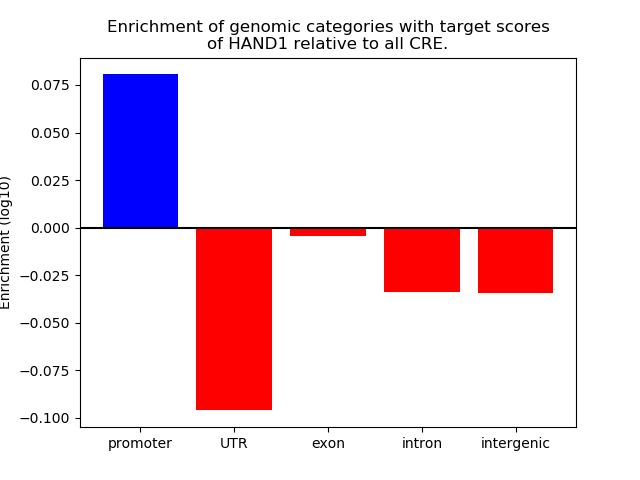
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_162602563_162603214 | 0.55 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
628 |
0.74 |
| chr12_57527040_57527217 | 0.55 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
1206 |
0.3 |
| chr1_29308973_29309136 | 0.49 |
ENSG00000206704 |
. |
3168 |
0.29 |
| chr20_53092813_53093232 | 0.47 |
DOK5 |
docking protein 5 |
765 |
0.8 |
| chr19_7581346_7582140 | 0.46 |
ZNF358 |
zinc finger protein 358 |
739 |
0.45 |
| chr11_114167265_114167463 | 0.45 |
NNMT |
nicotinamide N-methyltransferase |
721 |
0.72 |
| chr18_52988018_52988218 | 0.42 |
TCF4 |
transcription factor 4 |
1099 |
0.65 |
| chr11_118481086_118481679 | 0.40 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
3024 |
0.16 |
| chr1_161167282_161167469 | 0.40 |
ADAMTS4 |
ADAM metallopeptidase with thrombospondin type 1 motif, 4 |
1468 |
0.16 |
| chr1_219116097_219116279 | 0.38 |
LYPLAL1 |
lysophospholipase-like 1 |
230998 |
0.02 |
| chr17_78472143_78472537 | 0.37 |
NPTX1 |
neuronal pentraxin I |
21936 |
0.17 |
| chr13_20413212_20413363 | 0.36 |
RP11-61K9.2 |
|
2680 |
0.24 |
| chr13_33543686_33543957 | 0.36 |
KL |
klotho |
46386 |
0.17 |
| chr10_45471952_45472103 | 0.36 |
C10orf10 |
chromosome 10 open reading frame 10 |
2210 |
0.2 |
| chr2_169658994_169659163 | 0.35 |
NOSTRIN |
nitric oxide synthase trafficking |
29 |
0.98 |
| chr8_13131816_13132160 | 0.35 |
DLC1 |
deleted in liver cancer 1 |
2067 |
0.42 |
| chr16_51180371_51180536 | 0.34 |
AC009166.5 |
|
2697 |
0.32 |
| chr9_95508973_95509124 | 0.34 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
18046 |
0.19 |
| chr14_52117494_52117667 | 0.34 |
FRMD6 |
FERM domain containing 6 |
996 |
0.57 |
| chr8_55294582_55295235 | 0.32 |
ENSG00000244107 |
. |
38120 |
0.17 |
| chr3_188692889_188693197 | 0.32 |
TPRG1 |
tumor protein p63 regulated 1 |
28040 |
0.26 |
| chr18_12420846_12421147 | 0.32 |
SLMO1 |
slowmo homolog 1 (Drosophila) |
289 |
0.89 |
| chr14_52708086_52708298 | 0.32 |
PTGDR |
prostaglandin D2 receptor (DP) |
26239 |
0.22 |
| chr1_46379145_46379296 | 0.31 |
MAST2 |
microtubule associated serine/threonine kinase 2 |
40 |
0.98 |
| chr8_71580077_71580228 | 0.31 |
LACTB2 |
lactamase, beta 2 |
1240 |
0.33 |
| chr16_73809854_73810059 | 0.31 |
PSMD7 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
520717 |
0.0 |
| chr8_58716743_58716938 | 0.30 |
FAM110B |
family with sequence similarity 110, member B |
190273 |
0.03 |
| chr12_50478440_50478601 | 0.29 |
SMARCD1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
235 |
0.87 |
| chr17_29891996_29892164 | 0.29 |
ENSG00000221038 |
. |
1167 |
0.33 |
| chr7_2654317_2654708 | 0.29 |
TTYH3 |
tweety family member 3 |
17073 |
0.16 |
| chr6_3759558_3759709 | 0.29 |
PXDC1 |
PX domain containing 1 |
7373 |
0.21 |
| chr13_106567585_106567736 | 0.29 |
ENSG00000252550 |
. |
17668 |
0.3 |
| chr8_25026050_25026201 | 0.28 |
ENSG00000241811 |
. |
13110 |
0.23 |
| chr15_81073340_81073491 | 0.27 |
KIAA1199 |
KIAA1199 |
1703 |
0.42 |
| chr1_163039657_163039882 | 0.27 |
RGS4 |
regulator of G-protein signaling 4 |
618 |
0.82 |
| chr9_14925125_14925276 | 0.27 |
FREM1 |
FRAS1 related extracellular matrix 1 |
14207 |
0.26 |
| chr3_179659864_179660182 | 0.27 |
PEX5L |
peroxisomal biogenesis factor 5-like |
31017 |
0.18 |
| chr6_56707059_56707245 | 0.27 |
DST |
dystonin |
791 |
0.6 |
| chr16_65154176_65154454 | 0.26 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
1518 |
0.59 |
| chr7_134551561_134551712 | 0.26 |
CALD1 |
caldesmon 1 |
44 |
0.99 |
| chr11_36052910_36053061 | 0.26 |
ENSG00000263389 |
. |
21337 |
0.21 |
| chr3_44626439_44626876 | 0.26 |
ZNF660 |
zinc finger protein 660 |
201 |
0.58 |
| chr15_96878465_96878729 | 0.26 |
ENSG00000222651 |
. |
2107 |
0.24 |
| chr17_65516545_65516723 | 0.26 |
CTD-2653B5.1 |
|
3963 |
0.23 |
| chr22_32340974_32341439 | 0.25 |
C22orf24 |
chromosome 22 open reading frame 24 |
130 |
0.89 |
| chr16_72893250_72893401 | 0.25 |
ENSG00000251868 |
. |
37434 |
0.15 |
| chr8_117670708_117670893 | 0.25 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
97223 |
0.08 |
| chr17_78044749_78045392 | 0.25 |
CCDC40 |
coiled-coil domain containing 40 |
12346 |
0.15 |
| chr14_51944904_51945055 | 0.25 |
FRMD6 |
FERM domain containing 6 |
10876 |
0.24 |
| chr7_1163898_1164053 | 0.25 |
C7orf50 |
chromosome 7 open reading frame 50 |
13901 |
0.12 |
| chr2_101478621_101478772 | 0.25 |
AC092168.2 |
|
39573 |
0.16 |
| chr8_93029441_93029592 | 0.25 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
75 |
0.98 |
| chr10_104813584_104814284 | 0.25 |
NT5C2 |
5'-nucleotidase, cytosolic II |
40191 |
0.17 |
| chr16_64484459_64484610 | 0.24 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
609047 |
0.0 |
| chr5_152900712_152900863 | 0.24 |
GRIA1 |
glutamate receptor, ionotropic, AMPA 1 |
29055 |
0.26 |
| chr17_34040098_34040306 | 0.24 |
RASL10B |
RAS-like, family 10, member B |
18466 |
0.11 |
| chr12_72663874_72664300 | 0.24 |
ENSG00000236333 |
. |
1684 |
0.39 |
| chr17_66511128_66511617 | 0.24 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
108 |
0.97 |
| chr10_90711644_90712353 | 0.23 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
532 |
0.73 |
| chr14_30610639_30610790 | 0.23 |
PRKD1 |
protein kinase D1 |
50390 |
0.19 |
| chr18_56245541_56245920 | 0.23 |
RP11-126O1.2 |
|
21428 |
0.14 |
| chr5_112033189_112033368 | 0.23 |
APC |
adenomatous polyposis coli |
9917 |
0.26 |
| chr3_114790059_114790273 | 0.23 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
56 |
0.99 |
| chr10_52834791_52834942 | 0.23 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
932 |
0.63 |
| chr5_129241974_129242191 | 0.23 |
CTC-575N7.1 |
|
472 |
0.8 |
| chr17_39969274_39969651 | 0.23 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
279 |
0.68 |
| chr12_1710538_1710689 | 0.23 |
FBXL14 |
F-box and leucine-rich repeat protein 14 |
7282 |
0.21 |
| chr4_188093462_188093613 | 0.22 |
ENSG00000252382 |
. |
314927 |
0.01 |
| chr9_112888297_112888507 | 0.22 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
621 |
0.82 |
| chr7_99933726_99933950 | 0.22 |
PILRB |
paired immunoglobin-like type 2 receptor beta |
101 |
0.92 |
| chr10_95517973_95518124 | 0.22 |
LGI1 |
leucine-rich, glioma inactivated 1 |
370 |
0.9 |
| chr2_217364608_217364845 | 0.22 |
RPL37A |
ribosomal protein L37a |
863 |
0.45 |
| chr5_143584679_143584889 | 0.22 |
KCTD16 |
potassium channel tetramerization domain containing 16 |
56 |
0.98 |
| chr7_100200060_100200259 | 0.22 |
PCOLCE |
procollagen C-endopeptidase enhancer |
359 |
0.52 |
| chr7_102576116_102576267 | 0.22 |
LRRC17 |
leucine rich repeat containing 17 |
2514 |
0.27 |
| chr15_100782457_100782608 | 0.22 |
ADAMTS17 |
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
98809 |
0.07 |
| chr11_119292436_119293234 | 0.22 |
THY1 |
Thy-1 cell surface antigen |
437 |
0.74 |
| chr3_25980008_25980159 | 0.21 |
LINC00692 |
long intergenic non-protein coding RNA 692 |
64894 |
0.13 |
| chr19_29439287_29439438 | 0.21 |
ENSG00000238514 |
. |
17452 |
0.26 |
| chr1_45277728_45277914 | 0.21 |
BTBD19 |
BTB (POZ) domain containing 19 |
3626 |
0.09 |
| chr2_12860250_12860601 | 0.21 |
TRIB2 |
tribbles pseudokinase 2 |
2061 |
0.41 |
| chr9_132033008_132033468 | 0.21 |
ENSG00000220992 |
. |
40242 |
0.13 |
| chr1_215905303_215905454 | 0.21 |
ENSG00000202498 |
. |
101919 |
0.08 |
| chr22_30641304_30641455 | 0.21 |
RP1-102K2.8 |
|
605 |
0.52 |
| chr16_69309884_69310035 | 0.21 |
RP11-343C2.11 |
Uncharacterized protein |
23626 |
0.1 |
| chr7_3299969_3300120 | 0.21 |
SDK1 |
sidekick cell adhesion molecule 1 |
41036 |
0.16 |
| chr10_77731686_77731838 | 0.21 |
ENSG00000215921 |
. |
148226 |
0.04 |
| chr3_134125711_134125946 | 0.21 |
ENSG00000221313 |
. |
3154 |
0.25 |
| chr3_171581116_171581308 | 0.20 |
TMEM212 |
transmembrane protein 212 |
19906 |
0.18 |
| chr9_90873059_90873210 | 0.20 |
ENSG00000252299 |
. |
116050 |
0.06 |
| chr1_150245169_150245508 | 0.20 |
C1orf54 |
chromosome 1 open reading frame 54 |
155 |
0.89 |
| chr10_127350541_127350692 | 0.20 |
TEX36 |
testis expressed 36 |
21059 |
0.17 |
| chr1_110455050_110455346 | 0.20 |
CSF1 |
colony stimulating factor 1 (macrophage) |
1590 |
0.37 |
| chr3_196324835_196325124 | 0.20 |
FBXO45 |
F-box protein 45 |
29420 |
0.1 |
| chr21_17828537_17828688 | 0.20 |
ENSG00000207638 |
. |
82797 |
0.1 |
| chr13_24477531_24477691 | 0.20 |
C1QTNF9B |
C1q and tumor necrosis factor related protein 9B |
817 |
0.59 |
| chr10_97415335_97415893 | 0.20 |
ALDH18A1 |
aldehyde dehydrogenase 18 family, member A1 |
849 |
0.66 |
| chr15_48931516_48931667 | 0.20 |
FBN1 |
fibrillin 1 |
6327 |
0.29 |
| chr14_76168479_76168630 | 0.19 |
TTLL5 |
tubulin tyrosine ligase-like family, member 5 |
40933 |
0.14 |
| chr4_154059352_154059503 | 0.19 |
TRIM2 |
tripartite motif containing 2 |
14218 |
0.26 |
| chr1_5998344_5998611 | 0.19 |
ENSG00000266687 |
. |
44493 |
0.12 |
| chr6_2377522_2377693 | 0.19 |
ENSG00000266252 |
. |
32262 |
0.24 |
| chr12_109569450_109569601 | 0.19 |
ACACB |
acetyl-CoA carboxylase beta |
362 |
0.84 |
| chr9_98412715_98412927 | 0.19 |
DKFZP434H0512 |
Protein LOC100506667; Putative uncharacterized protein DKFZp434H0512 |
121784 |
0.05 |
| chr18_53069042_53069242 | 0.19 |
TCF4 |
transcription factor 4 |
60 |
0.98 |
| chr18_22813505_22813656 | 0.19 |
ZNF521 |
zinc finger protein 521 |
2086 |
0.46 |
| chr3_123601954_123602216 | 0.19 |
MYLK |
myosin light chain kinase |
1064 |
0.59 |
| chr5_80251688_80251866 | 0.19 |
CTC-459I6.1 |
|
2978 |
0.3 |
| chr10_27909508_27909724 | 0.19 |
RAB18 |
RAB18, member RAS oncogene family |
116068 |
0.06 |
| chr10_126223452_126223603 | 0.19 |
LHPP |
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
73115 |
0.09 |
| chr7_102489714_102489865 | 0.19 |
ENSG00000238324 |
. |
8108 |
0.18 |
| chr11_2720232_2720685 | 0.19 |
KCNQ1OT1 |
KCNQ1 opposite strand/antisense transcript 1 (non-protein coding) |
766 |
0.73 |
| chr3_194409728_194410184 | 0.19 |
FAM43A |
family with sequence similarity 43, member A |
3334 |
0.19 |
| chr1_153515695_153515846 | 0.19 |
S100A5 |
S100 calcium binding protein A5 |
1529 |
0.18 |
| chr2_130691083_130691234 | 0.18 |
AC079776.2 |
|
7797 |
0.21 |
| chr11_87151692_87151843 | 0.18 |
ENSG00000223015 |
. |
165908 |
0.04 |
| chr15_50139702_50139853 | 0.18 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
29115 |
0.19 |
| chr5_176942929_176943080 | 0.18 |
DDX41 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 |
913 |
0.39 |
| chr3_194900793_194900985 | 0.18 |
XXYLT1 |
xyloside xylosyltransferase 1 |
4487 |
0.18 |
| chr1_201624831_201624982 | 0.18 |
NAV1 |
neuron navigator 1 |
7456 |
0.16 |
| chr8_102550339_102550506 | 0.18 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
45436 |
0.15 |
| chr7_4707772_4707923 | 0.18 |
FOXK1 |
forkhead box K1 |
14093 |
0.21 |
| chr11_73023790_73024080 | 0.18 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
1325 |
0.34 |
| chr12_13133119_13133304 | 0.18 |
RP11-392P7.6 |
|
440 |
0.74 |
| chr2_192501340_192501491 | 0.18 |
NABP1 |
nucleic acid binding protein 1 |
41447 |
0.2 |
| chr12_81472109_81472459 | 0.18 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
448 |
0.87 |
| chr12_20628731_20628918 | 0.18 |
RP11-284H19.1 |
|
105628 |
0.07 |
| chr9_35848269_35848420 | 0.18 |
TMEM8B |
transmembrane protein 8B |
18831 |
0.09 |
| chr6_169990007_169990169 | 0.18 |
WDR27 |
WD repeat domain 27 |
70700 |
0.1 |
| chr21_44061431_44061649 | 0.17 |
AP001626.2 |
|
9814 |
0.18 |
| chr1_163055295_163055446 | 0.17 |
RGS4 |
regulator of G-protein signaling 4 |
13434 |
0.25 |
| chr2_152919196_152919405 | 0.17 |
AC079790.2 |
|
35542 |
0.18 |
| chr19_39647674_39647825 | 0.17 |
PAK4 |
p21 protein (Cdc42/Rac)-activated kinase 4 |
407 |
0.78 |
| chrX_36975622_36976198 | 0.17 |
FAM47C |
family with sequence similarity 47, member C |
50522 |
0.19 |
| chr17_76858323_76858500 | 0.17 |
RP11-323N12.5 |
|
6851 |
0.15 |
| chr13_108472165_108472316 | 0.17 |
FAM155A-IT1 |
FAM155A intronic transcript 1 (non-protein coding) |
15559 |
0.26 |
| chr6_34624121_34624272 | 0.17 |
C6orf106 |
chromosome 6 open reading frame 106 |
15537 |
0.14 |
| chr9_4294012_4294168 | 0.17 |
GLIS3 |
GLIS family zinc finger 3 |
4406 |
0.24 |
| chr10_81203831_81204005 | 0.17 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
57 |
0.98 |
| chr5_121409447_121409998 | 0.17 |
LOX |
lysyl oxidase |
4258 |
0.27 |
| chr17_45982661_45983102 | 0.17 |
SP2 |
Sp2 transcription factor |
9365 |
0.1 |
| chr20_2449616_2449767 | 0.17 |
SNRPB |
small nuclear ribonucleoprotein polypeptides B and B1 |
1740 |
0.28 |
| chr12_117042641_117042795 | 0.17 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
29062 |
0.22 |
| chr2_207082896_207083047 | 0.17 |
GPR1 |
G protein-coupled receptor 1 |
200 |
0.91 |
| chr18_43573899_43574073 | 0.17 |
ENSG00000222179 |
. |
4075 |
0.23 |
| chr8_105337886_105338037 | 0.17 |
DCSTAMP |
dendrocyte expressed seven transmembrane protein |
14093 |
0.23 |
| chr3_114098944_114099095 | 0.16 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
3469 |
0.25 |
| chr10_65227518_65227669 | 0.16 |
JMJD1C |
jumonji domain containing 1C |
1871 |
0.34 |
| chr17_7153794_7154016 | 0.16 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
700 |
0.29 |
| chr11_46311932_46312209 | 0.16 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
4607 |
0.2 |
| chr13_67708586_67709053 | 0.16 |
PCDH9 |
protocadherin 9 |
93753 |
0.09 |
| chr7_140049305_140049502 | 0.16 |
SLC37A3 |
solute carrier family 37, member 3 |
917 |
0.52 |
| chr1_29450572_29451688 | 0.16 |
TMEM200B |
transmembrane protein 200B |
683 |
0.7 |
| chr2_236525231_236525382 | 0.16 |
ENSG00000221704 |
. |
44731 |
0.16 |
| chr2_136578774_136578925 | 0.16 |
AC011893.3 |
|
1088 |
0.47 |
| chr11_128353135_128353286 | 0.16 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
22079 |
0.23 |
| chr13_28711690_28712227 | 0.16 |
PAN3-AS1 |
PAN3 antisense RNA 1 |
372 |
0.68 |
| chr4_127551270_127551421 | 0.16 |
ENSG00000199862 |
. |
398029 |
0.01 |
| chr17_70451846_70451997 | 0.16 |
ENSG00000200783 |
. |
208370 |
0.02 |
| chr14_79032841_79033001 | 0.16 |
NRXN3 |
neurexin 3 |
48354 |
0.15 |
| chr3_156873305_156873620 | 0.16 |
ENSG00000201778 |
. |
2125 |
0.24 |
| chr6_27125358_27125562 | 0.16 |
ENSG00000265565 |
. |
10055 |
0.11 |
| chr17_15522812_15523031 | 0.16 |
CDRT1 |
CMT1A duplicated region transcript 1 |
95 |
0.95 |
| chr10_29771962_29772274 | 0.16 |
SVIL |
supervillin |
13329 |
0.23 |
| chr19_16436739_16437476 | 0.16 |
KLF2 |
Kruppel-like factor 2 |
1456 |
0.3 |
| chr10_112431738_112432014 | 0.16 |
RBM20 |
RNA binding motif protein 20 |
27721 |
0.14 |
| chr13_21808635_21808786 | 0.16 |
MRP63 |
mitochondrial ribosomal protein 63 |
57926 |
0.09 |
| chr13_68315780_68315931 | 0.16 |
PCDH9 |
protocadherin 9 |
511387 |
0.0 |
| chr5_54898438_54898589 | 0.16 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
67635 |
0.1 |
| chr17_59338039_59338190 | 0.16 |
RP11-332H18.3 |
|
122032 |
0.04 |
| chr5_42943877_42944134 | 0.16 |
SEPP1 |
selenoprotein P, plasma, 1 |
56511 |
0.11 |
| chr11_131852297_131852448 | 0.16 |
RP11-697E14.2 |
|
2183 |
0.42 |
| chr8_22133429_22133580 | 0.16 |
PIWIL2 |
piwi-like RNA-mediated gene silencing 2 |
424 |
0.64 |
| chr1_224363177_224363414 | 0.16 |
DEGS1 |
delta(4)-desaturase, sphingolipid 1 |
163 |
0.94 |
| chr12_16505567_16505718 | 0.16 |
MGST1 |
microsomal glutathione S-transferase 1 |
709 |
0.77 |
| chr2_33516384_33516581 | 0.16 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
233 |
0.94 |
| chr16_82769509_82769673 | 0.15 |
RP11-22H5.2 |
|
37333 |
0.19 |
| chr11_130545351_130545502 | 0.15 |
C11orf44 |
chromosome 11 open reading frame 44 |
2575 |
0.42 |
| chr2_25474203_25475088 | 0.15 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
535 |
0.81 |
| chr1_201618776_201619150 | 0.15 |
NAV1 |
neuron navigator 1 |
1513 |
0.35 |
| chr10_127860039_127860205 | 0.15 |
ENSG00000222740 |
. |
25971 |
0.24 |
| chr10_103113133_103113457 | 0.15 |
BTRC |
beta-transducin repeat containing E3 ubiquitin protein ligase |
525 |
0.74 |
| chr17_13018053_13018229 | 0.15 |
ELAC2 |
elaC ribonuclease Z 2 |
96637 |
0.08 |
| chr15_76004107_76004336 | 0.15 |
CSPG4 |
chondroitin sulfate proteoglycan 4 |
968 |
0.37 |
| chr15_39881812_39881998 | 0.15 |
CTD-2033D15.1 |
|
4527 |
0.21 |
| chr16_30420644_30420806 | 0.15 |
ZNF771 |
zinc finger protein 771 |
1373 |
0.2 |
| chr15_78171702_78171998 | 0.15 |
CSPG4P13 |
chondroitin sulfate proteoglycan 4 pseudogene 13 |
15176 |
0.15 |
| chr2_225795288_225795439 | 0.15 |
DOCK10 |
dedicator of cytokinesis 10 |
16419 |
0.28 |
| chr6_45705656_45705807 | 0.15 |
ENSG00000252738 |
. |
91890 |
0.09 |
| chr8_70406794_70407007 | 0.15 |
SULF1 |
sulfatase 1 |
1872 |
0.5 |
| chr11_46302791_46303236 | 0.15 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
3785 |
0.21 |
| chr5_137225208_137225431 | 0.15 |
PKD2L2 |
polycystic kidney disease 2-like 2 |
161 |
0.67 |
| chr10_88159807_88161210 | 0.15 |
GRID1 |
glutamate receptor, ionotropic, delta 1 |
34273 |
0.17 |
| chr6_75992353_75992507 | 0.15 |
TMEM30A |
transmembrane protein 30A |
1597 |
0.4 |
| chr7_150942822_150943143 | 0.15 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
3077 |
0.12 |
| chr6_137635380_137635531 | 0.15 |
IFNGR1 |
interferon gamma receptor 1 |
94869 |
0.08 |
| chr2_113403985_113404215 | 0.15 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
666 |
0.72 |
| chr20_21962515_21962666 | 0.15 |
PAX1 |
paired box 1 |
275729 |
0.02 |
| chr7_25654532_25654683 | 0.15 |
ENSG00000222101 |
. |
44319 |
0.21 |
| chr12_66220556_66220804 | 0.15 |
HMGA2 |
high mobility group AT-hook 2 |
1777 |
0.39 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.3 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.2 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.2 | GO:0006853 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.0 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.4 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.2 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.2 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.0 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.3 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.0 | 0.0 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:2001234 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.0 | GO:0045978 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.0 | 0.3 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.0 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0043631 | RNA polyadenylation(GO:0043631) |
| 0.0 | 0.0 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.0 | GO:0055094 | response to lipoprotein particle(GO:0055094) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.0 | GO:0002913 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.0 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.1 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0061042 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.0 | GO:0071548 | response to dexamethasone(GO:0071548) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.3 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.0 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0047115 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.0 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |