Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
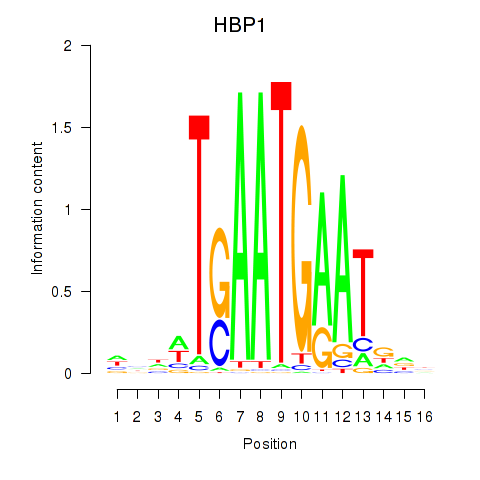
Results for HBP1
Z-value: 1.13
Transcription factors associated with HBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HBP1
|
ENSG00000105856.9 | HMG-box transcription factor 1 |
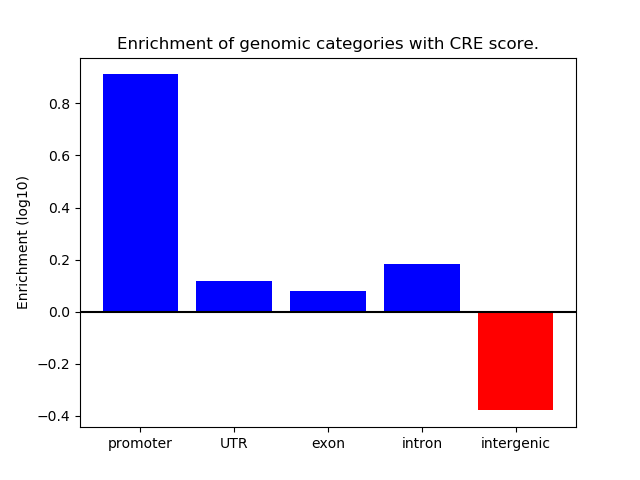
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_106874250_106874401 | HBP1 | 38032 | 0.151723 | 0.39 | 3.0e-01 | Click! |
| chr7_106808610_106808963 | HBP1 | 620 | 0.749796 | 0.34 | 3.7e-01 | Click! |
| chr7_106810018_106810657 | HBP1 | 119 | 0.968484 | 0.28 | 4.6e-01 | Click! |
| chr7_106807796_106808590 | HBP1 | 1213 | 0.499467 | 0.27 | 4.8e-01 | Click! |
| chr7_106874818_106874969 | HBP1 | 38600 | 0.150442 | 0.26 | 5.1e-01 | Click! |
Activity of the HBP1 motif across conditions
Conditions sorted by the z-value of the HBP1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
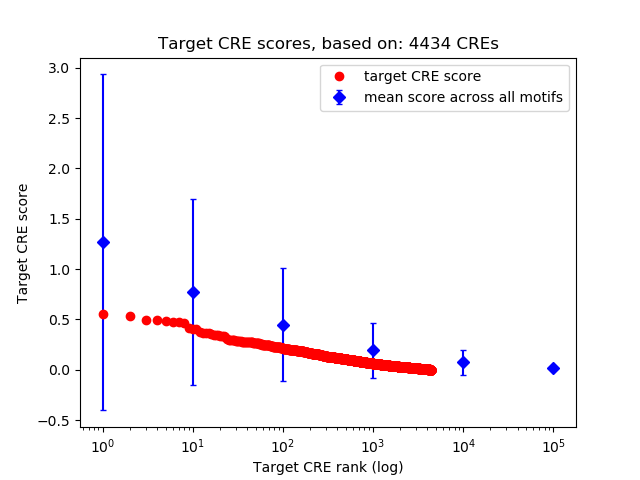
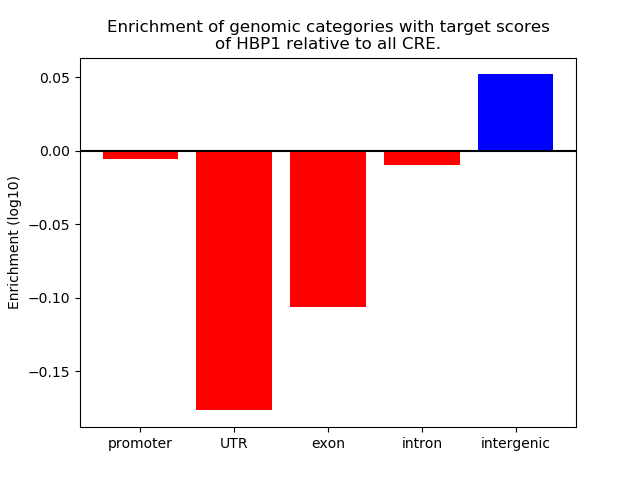
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_163171316_163171467 | 0.56 |
IFIH1 |
interferon induced with helicase C domain 1 |
3803 |
0.25 |
| chr19_49661333_49661989 | 0.53 |
TRPM4 |
transient receptor potential cation channel, subfamily M, member 4 |
562 |
0.58 |
| chr9_684288_685148 | 0.50 |
RP11-130C19.3 |
|
837 |
0.67 |
| chr18_9648789_9648940 | 0.50 |
ENSG00000212572 |
. |
30794 |
0.14 |
| chr3_105120211_105120362 | 0.49 |
ALCAM |
activated leukocyte cell adhesion molecule |
34099 |
0.25 |
| chr16_11295551_11295900 | 0.47 |
RMI2 |
RecQ mediated genome instability 2 |
47781 |
0.08 |
| chr7_194025_194290 | 0.47 |
AC093627.12 |
|
23 |
0.96 |
| chr2_113593759_113594336 | 0.46 |
IL1B |
interleukin 1, beta |
36 |
0.97 |
| chr17_74684871_74685203 | 0.42 |
MXRA7 |
matrix-remodelling associated 7 |
68 |
0.94 |
| chr14_78602062_78602325 | 0.41 |
ENSG00000199454 |
. |
42071 |
0.19 |
| chr4_114899439_114899761 | 0.41 |
ARSJ |
arylsulfatase family, member J |
552 |
0.84 |
| chr3_157154723_157155399 | 0.37 |
PTX3 |
pentraxin 3, long |
483 |
0.86 |
| chr18_47089345_47089666 | 0.37 |
LIPG |
lipase, endothelial |
790 |
0.6 |
| chr6_154675778_154676172 | 0.37 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
1894 |
0.48 |
| chr20_49638752_49638903 | 0.37 |
KCNG1 |
potassium voltage-gated channel, subfamily G, member 1 |
790 |
0.62 |
| chr7_155678768_155678919 | 0.35 |
ENSG00000221113 |
. |
14258 |
0.24 |
| chr9_80455109_80455260 | 0.35 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
17269 |
0.29 |
| chr18_7116438_7116788 | 0.35 |
LAMA1 |
laminin, alpha 1 |
1200 |
0.55 |
| chr11_65083293_65083503 | 0.34 |
CDC42EP2 |
CDC42 effector protein (Rho GTPase binding) 2 |
1109 |
0.37 |
| chr2_98703049_98703523 | 0.34 |
VWA3B |
von Willebrand factor A domain containing 3B |
309 |
0.94 |
| chr2_71115848_71116159 | 0.33 |
VAX2 |
ventral anterior homeobox 2 |
11717 |
0.13 |
| chr9_132168981_132169132 | 0.33 |
ENSG00000242281 |
. |
36316 |
0.16 |
| chr5_14265991_14266171 | 0.33 |
TRIO |
trio Rho guanine nucleotide exchange factor |
25005 |
0.28 |
| chr3_5055062_5055354 | 0.30 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
33562 |
0.15 |
| chr6_24743062_24743323 | 0.30 |
C6orf62 |
chromosome 6 open reading frame 62 |
22128 |
0.12 |
| chr8_9472125_9472276 | 0.30 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
39683 |
0.18 |
| chr10_95239483_95239707 | 0.30 |
MYOF |
myoferlin |
2356 |
0.29 |
| chr2_26624476_26625174 | 0.29 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
41 |
0.98 |
| chr3_11755297_11755448 | 0.29 |
VGLL4 |
vestigial like 4 (Drosophila) |
3361 |
0.34 |
| chr7_93694115_93694266 | 0.29 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
60496 |
0.13 |
| chr13_76217437_76217800 | 0.29 |
LMO7 |
LIM domain 7 |
7159 |
0.18 |
| chr7_113996777_113996928 | 0.28 |
FOXP2 |
forkhead box P2 |
57477 |
0.15 |
| chr5_62219537_62219688 | 0.28 |
LRRC70 |
leucine rich repeat containing 70 |
344910 |
0.01 |
| chr9_111251929_111252080 | 0.28 |
ENSG00000222512 |
. |
130795 |
0.06 |
| chr18_52274836_52275204 | 0.28 |
DYNAP |
dynactin associated protein |
16630 |
0.28 |
| chr10_21688030_21688514 | 0.28 |
ENSG00000207264 |
. |
77382 |
0.08 |
| chr22_38139385_38139536 | 0.28 |
TRIOBP |
TRIO and F-actin binding protein |
2775 |
0.17 |
| chr21_39288370_39288729 | 0.28 |
KCNJ6 |
potassium inwardly-rectifying channel, subfamily J, member 6 |
200 |
0.96 |
| chr17_7944787_7945045 | 0.28 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
2442 |
0.16 |
| chr1_85215722_85215873 | 0.27 |
ENSG00000251899 |
. |
43085 |
0.15 |
| chr4_85886471_85886934 | 0.27 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
801 |
0.75 |
| chr8_48770878_48771029 | 0.27 |
ENSG00000211562 |
. |
31666 |
0.17 |
| chr9_12859535_12860149 | 0.27 |
ENSG00000222658 |
. |
25478 |
0.2 |
| chr18_45494721_45494993 | 0.27 |
SMAD2 |
SMAD family member 2 |
37342 |
0.19 |
| chr2_213793817_213793968 | 0.27 |
ENSG00000266354 |
. |
2911 |
0.39 |
| chr15_34500911_34501100 | 0.27 |
KATNBL1 |
katanin p80 subunit B-like 1 |
1207 |
0.43 |
| chr5_145318270_145318548 | 0.27 |
SH3RF2 |
SH3 domain containing ring finger 2 |
969 |
0.66 |
| chr1_153584096_153584410 | 0.27 |
S100A16 |
S100 calcium binding protein A16 |
1306 |
0.22 |
| chr9_39816869_39817104 | 0.27 |
SPATA31A2 |
SPATA31 subfamily A, member 2 |
67989 |
0.13 |
| chr6_141587373_141587724 | 0.27 |
ENSG00000222764 |
. |
220001 |
0.02 |
| chr19_8633826_8634145 | 0.27 |
MYO1F |
myosin IF |
8337 |
0.12 |
| chr17_37301005_37301156 | 0.27 |
PLXDC1 |
plexin domain containing 1 |
6822 |
0.14 |
| chr1_16647953_16648232 | 0.27 |
FBXO42 |
F-box protein 42 |
30857 |
0.12 |
| chr9_39287616_39287852 | 0.26 |
CNTNAP3 |
contactin associated protein-like 3 |
358 |
0.92 |
| chr5_177541606_177541785 | 0.26 |
N4BP3 |
NEDD4 binding protein 3 |
1251 |
0.43 |
| chr7_96971795_96971946 | 0.26 |
ENSG00000221192 |
. |
50962 |
0.18 |
| chr5_141706853_141707016 | 0.26 |
AC005592.2 |
|
2071 |
0.33 |
| chr4_153770751_153770902 | 0.26 |
ENSG00000252375 |
. |
51292 |
0.13 |
| chr6_3238151_3238302 | 0.25 |
PSMG4 |
proteasome (prosome, macropain) assembly chaperone 4 |
6185 |
0.19 |
| chr17_39196389_39196540 | 0.25 |
KRTAP1-1 |
keratin associated protein 1-1 |
1249 |
0.19 |
| chr1_16125825_16125998 | 0.25 |
FBLIM1 |
filamin binding LIM protein 1 |
34463 |
0.09 |
| chr3_174485599_174485896 | 0.25 |
NAALADL2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
91323 |
0.1 |
| chr20_11871067_11871503 | 0.25 |
BTBD3 |
BTB (POZ) domain containing 3 |
86 |
0.98 |
| chr20_38245634_38245785 | 0.25 |
ENSG00000241840 |
. |
264307 |
0.02 |
| chr5_156921339_156921490 | 0.25 |
ADAM19 |
ADAM metallopeptidase domain 19 |
8416 |
0.17 |
| chr4_100814684_100815546 | 0.25 |
LAMTOR3 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
418 |
0.86 |
| chr20_14900281_14900452 | 0.24 |
MACROD2-AS1 |
MACROD2 antisense RNA 1 |
9766 |
0.23 |
| chr2_37589065_37589216 | 0.24 |
QPCT |
glutaminyl-peptide cyclotransferase |
6284 |
0.2 |
| chr19_45147190_45148055 | 0.24 |
PVR |
poliovirus receptor |
225 |
0.9 |
| chr2_161265827_161265978 | 0.24 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
782 |
0.57 |
| chr4_40843301_40843452 | 0.24 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
1157 |
0.46 |
| chr1_97025244_97025565 | 0.24 |
ENSG00000241992 |
. |
23361 |
0.27 |
| chr1_38941740_38942113 | 0.24 |
ENSG00000200796 |
. |
79595 |
0.11 |
| chr1_116470870_116471021 | 0.24 |
SLC22A15 |
solute carrier family 22, member 15 |
48174 |
0.15 |
| chr7_41740779_41741576 | 0.24 |
INHBA |
inhibin, beta A |
970 |
0.56 |
| chr15_70393365_70393548 | 0.23 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
2941 |
0.32 |
| chr3_188599632_188599783 | 0.23 |
TPRG1 |
tumor protein p63 regulated 1 |
65296 |
0.14 |
| chr13_104713968_104714119 | 0.23 |
SLC10A2 |
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
994847 |
0.0 |
| chr9_137393023_137393337 | 0.23 |
RXRA |
retinoid X receptor, alpha |
94752 |
0.07 |
| chr6_108886554_108887901 | 0.23 |
FOXO3 |
forkhead box O3 |
5158 |
0.32 |
| chr4_177931083_177931234 | 0.23 |
ENSG00000222859 |
. |
171438 |
0.03 |
| chr22_44844083_44844234 | 0.23 |
LDOC1L |
leucine zipper, down-regulated in cancer 1-like |
50020 |
0.16 |
| chr9_80539592_80539743 | 0.23 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
101752 |
0.08 |
| chr14_54430258_54430823 | 0.23 |
BMP4 |
bone morphogenetic protein 4 |
5061 |
0.28 |
| chr6_77745416_77745567 | 0.23 |
HTR1B |
5-hydroxytryptamine (serotonin) receptor 1B, G protein-coupled |
427999 |
0.01 |
| chr12_78334342_78334902 | 0.22 |
NAV3 |
neuron navigator 3 |
25434 |
0.27 |
| chr1_178067294_178067727 | 0.22 |
RASAL2 |
RAS protein activator like 2 |
4234 |
0.36 |
| chr9_43685064_43685767 | 0.22 |
CNTNAP3B |
contactin associated protein-like 3B |
505 |
0.86 |
| chr3_14644766_14644917 | 0.22 |
AC090952.5 |
|
46098 |
0.13 |
| chr1_94071277_94071503 | 0.22 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
8264 |
0.21 |
| chr19_18287388_18287660 | 0.22 |
IFI30 |
interferon, gamma-inducible protein 30 |
3047 |
0.14 |
| chr5_161768104_161768255 | 0.22 |
ENSG00000201474 |
. |
136718 |
0.05 |
| chr7_7223417_7223573 | 0.22 |
C1GALT1 |
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
1249 |
0.52 |
| chr1_85929812_85930054 | 0.22 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
334 |
0.91 |
| chr18_10787530_10787681 | 0.22 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
465 |
0.88 |
| chr1_179109732_179109883 | 0.22 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
2372 |
0.31 |
| chr4_152462150_152462592 | 0.22 |
FAM160A1 |
family with sequence similarity 160, member A1 |
8207 |
0.21 |
| chr3_128712183_128712559 | 0.22 |
KIAA1257 |
KIAA1257 |
548 |
0.71 |
| chr9_41432727_41433091 | 0.22 |
SPATA31A5 |
SPATA31 subfamily A, member 5 |
67770 |
0.13 |
| chr2_161253806_161254194 | 0.21 |
ENSG00000252465 |
. |
521 |
0.8 |
| chr2_36589107_36589258 | 0.21 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
5568 |
0.33 |
| chrX_143576959_143577110 | 0.21 |
ENSG00000201912 |
. |
561594 |
0.0 |
| chr2_219269994_219270145 | 0.21 |
ENSG00000199121 |
. |
2700 |
0.14 |
| chr15_72489601_72489809 | 0.21 |
GRAMD2 |
GRAM domain containing 2 |
421 |
0.81 |
| chr16_10174074_10174418 | 0.21 |
RP11-895K13.2 |
|
46705 |
0.18 |
| chr6_17394327_17394802 | 0.21 |
CAP2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
621 |
0.83 |
| chr8_134742646_134742797 | 0.21 |
ENSG00000212273 |
. |
85753 |
0.11 |
| chr7_51101841_51102176 | 0.21 |
RP4-724E13.2 |
|
16848 |
0.28 |
| chr20_60163701_60163894 | 0.21 |
CDH4 |
cadherin 4, type 1, R-cadherin (retinal) |
11037 |
0.3 |
| chr3_47584632_47584783 | 0.21 |
ENSG00000266443 |
. |
3117 |
0.18 |
| chr3_31931620_31931771 | 0.21 |
OSBPL10 |
oxysterol binding protein-like 10 |
60098 |
0.13 |
| chr20_17553764_17553929 | 0.20 |
DSTN |
destrin (actin depolymerizing factor) |
3119 |
0.21 |
| chr10_126752255_126752406 | 0.20 |
ENSG00000264572 |
. |
30891 |
0.18 |
| chr19_51874762_51874926 | 0.20 |
NKG7 |
natural killer cell group 7 sequence |
813 |
0.34 |
| chr20_47404517_47404913 | 0.20 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
39705 |
0.18 |
| chr8_23399467_23399639 | 0.20 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
12980 |
0.16 |
| chr6_83107092_83107769 | 0.20 |
TPBG |
trophoblast glycoprotein |
33469 |
0.24 |
| chr1_61550208_61550359 | 0.20 |
ENSG00000263380 |
. |
1081 |
0.48 |
| chr1_155196952_155197285 | 0.20 |
GBAP1 |
glucosidase, beta, acid pseudogene 1 |
96 |
0.89 |
| chr2_153028407_153028558 | 0.20 |
STAM2 |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
4024 |
0.3 |
| chr20_50543681_50543832 | 0.20 |
ENSG00000263659 |
. |
13616 |
0.19 |
| chr3_185815383_185815746 | 0.20 |
ETV5 |
ets variant 5 |
10769 |
0.22 |
| chr22_30609203_30609443 | 0.20 |
RP3-438O4.4 |
|
6225 |
0.13 |
| chr8_25041394_25041951 | 0.20 |
DOCK5 |
dedicator of cytokinesis 5 |
566 |
0.78 |
| chr3_168744436_168744695 | 0.20 |
MECOM |
MDS1 and EVI1 complex locus |
101257 |
0.09 |
| chr7_130594661_130594812 | 0.20 |
ENSG00000226380 |
. |
32438 |
0.2 |
| chr10_4719979_4720356 | 0.20 |
AKR1E2 |
aldo-keto reductase family 1, member E2 |
108654 |
0.07 |
| chr11_76535039_76535190 | 0.20 |
RP11-21L23.2 |
|
23706 |
0.13 |
| chr12_101416214_101416365 | 0.20 |
RP11-350G24.1 |
|
16583 |
0.22 |
| chr11_119630384_119630535 | 0.20 |
CTD-2523D13.2 |
|
30166 |
0.17 |
| chr2_220527194_220527345 | 0.20 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
31417 |
0.11 |
| chr6_56212766_56212917 | 0.20 |
RP3-445N2.1 |
|
15926 |
0.24 |
| chr7_34026746_34026942 | 0.20 |
BMPER |
BMP binding endothelial regulator |
81699 |
0.11 |
| chr7_18549761_18549974 | 0.20 |
HDAC9 |
histone deacetylase 9 |
931 |
0.71 |
| chr16_53243110_53243293 | 0.20 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
838 |
0.65 |
| chr2_150998838_150998989 | 0.19 |
RND3 |
Rho family GTPase 3 |
342983 |
0.01 |
| chr3_175150553_175150931 | 0.19 |
ENSG00000199792 |
. |
35605 |
0.2 |
| chr20_57602733_57603209 | 0.19 |
ATP5E |
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
4376 |
0.17 |
| chr2_112456971_112457256 | 0.19 |
ENSG00000266063 |
. |
71598 |
0.12 |
| chr9_68409245_68409396 | 0.19 |
ENSG00000207277 |
. |
396 |
0.88 |
| chr2_237780655_237780806 | 0.19 |
ENSG00000202341 |
. |
143218 |
0.04 |
| chr9_133822496_133822647 | 0.19 |
FIBCD1 |
fibrinogen C domain containing 1 |
8044 |
0.18 |
| chr9_116384910_116385169 | 0.19 |
RGS3 |
regulator of G-protein signaling 3 |
29273 |
0.19 |
| chr21_36216765_36216916 | 0.19 |
RUNX1 |
runt-related transcription factor 1 |
42640 |
0.19 |
| chr2_74374273_74375067 | 0.19 |
BOLA3 |
bolA family member 3 |
362 |
0.54 |
| chr3_32464050_32464201 | 0.19 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
30594 |
0.19 |
| chr22_22216534_22216685 | 0.19 |
MAPK1 |
mitogen-activated protein kinase 1 |
5121 |
0.18 |
| chr4_105890617_105890768 | 0.19 |
ENSG00000251906 |
. |
5127 |
0.33 |
| chr11_66823772_66824268 | 0.19 |
RHOD |
ras homolog family member D |
269 |
0.9 |
| chr11_46935807_46936022 | 0.19 |
LRP4 |
low density lipoprotein receptor-related protein 4 |
4259 |
0.21 |
| chr15_56351412_56351563 | 0.19 |
ENSG00000239703 |
. |
29308 |
0.2 |
| chr7_56101285_56102227 | 0.19 |
PSPH |
phosphoserine phosphatase |
93 |
0.95 |
| chr1_26680354_26680505 | 0.19 |
AIM1L |
absent in melanoma 1-like |
192 |
0.9 |
| chr3_167814440_167814591 | 0.19 |
GOLIM4 |
golgi integral membrane protein 4 |
752 |
0.79 |
| chr13_31272137_31272539 | 0.19 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
37307 |
0.19 |
| chr8_11721948_11722219 | 0.19 |
CTSB |
cathepsin B |
2106 |
0.27 |
| chr17_79007421_79007691 | 0.19 |
BAIAP2 |
BAI1-associated protein 2 |
1394 |
0.31 |
| chr4_105982764_105983024 | 0.19 |
ENSG00000252136 |
. |
44250 |
0.17 |
| chr5_175116482_175116633 | 0.18 |
HRH2 |
histamine receptor H2 |
8093 |
0.21 |
| chr9_123476537_123476776 | 0.18 |
MEGF9 |
multiple EGF-like-domains 9 |
44 |
0.98 |
| chr17_62257398_62257549 | 0.18 |
TEX2 |
testis expressed 2 |
32606 |
0.13 |
| chr6_148513736_148514010 | 0.18 |
SASH1 |
SAM and SH3 domain containing 1 |
79567 |
0.1 |
| chr3_156346028_156346179 | 0.18 |
TIPARP-AS1 |
TIPARP antisense RNA 1 |
44556 |
0.14 |
| chr8_22932847_22932998 | 0.18 |
RP11-875O11.3 |
|
921 |
0.44 |
| chr8_107059298_107059449 | 0.18 |
ENSG00000251003 |
. |
13298 |
0.29 |
| chr3_48470007_48470158 | 0.18 |
PLXNB1 |
plexin B1 |
790 |
0.43 |
| chr22_18255963_18256689 | 0.18 |
BID |
BH3 interacting domain death agonist |
456 |
0.82 |
| chr2_38144201_38144470 | 0.18 |
RMDN2 |
regulator of microtubule dynamics 2 |
5995 |
0.3 |
| chr8_119121667_119121966 | 0.18 |
EXT1 |
exostosin glycosyltransferase 1 |
837 |
0.77 |
| chr2_71127433_71128165 | 0.18 |
VAX2 |
ventral anterior homeobox 2 |
79 |
0.96 |
| chr4_77131865_77132211 | 0.18 |
SCARB2 |
scavenger receptor class B, member 2 |
2718 |
0.22 |
| chr12_81489356_81489507 | 0.18 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
17595 |
0.21 |
| chr1_94711175_94711488 | 0.18 |
ARHGAP29 |
Rho GTPase activating protein 29 |
8142 |
0.25 |
| chr8_128699254_128699540 | 0.18 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
48283 |
0.18 |
| chr1_186156970_186157465 | 0.18 |
GS1-174L6.4 |
|
10783 |
0.22 |
| chr2_113998091_113998347 | 0.18 |
RP11-65I12.1 |
|
88 |
0.94 |
| chr7_143072173_143072546 | 0.18 |
AC093673.5 |
|
5229 |
0.12 |
| chr6_27205685_27206711 | 0.18 |
PRSS16 |
protease, serine, 16 (thymus) |
9282 |
0.19 |
| chr5_133802242_133802478 | 0.18 |
ENSG00000207222 |
. |
33615 |
0.12 |
| chr9_123657022_123657322 | 0.18 |
PHF19 |
PHD finger protein 19 |
17566 |
0.17 |
| chr14_94856045_94856196 | 0.17 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
831 |
0.62 |
| chr1_182993880_182994112 | 0.17 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
1401 |
0.44 |
| chr10_7806287_7806438 | 0.17 |
KIN |
KIN, antigenic determinant of recA protein homolog (mouse) |
23582 |
0.17 |
| chr1_178064247_178064419 | 0.17 |
RASAL2 |
RAS protein activator like 2 |
1057 |
0.69 |
| chr10_52750817_52751095 | 0.17 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
11 |
0.99 |
| chr16_28228037_28228188 | 0.17 |
XPO6 |
exportin 6 |
4871 |
0.21 |
| chr13_77148711_77148862 | 0.17 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
311739 |
0.01 |
| chr7_41739624_41740047 | 0.17 |
INHBA |
inhibin, beta A |
372 |
0.87 |
| chr10_23734346_23734497 | 0.17 |
OTUD1 |
OTU domain containing 1 |
6223 |
0.23 |
| chr13_40974473_40974748 | 0.17 |
ENSG00000252812 |
. |
6967 |
0.31 |
| chr10_79470838_79471281 | 0.17 |
ENSG00000199664 |
. |
65754 |
0.1 |
| chr2_851043_851194 | 0.17 |
AC113607.3 |
|
65692 |
0.12 |
| chr10_37761163_37761314 | 0.17 |
MTRNR2L7 |
MT-RNR2-like 7 |
130621 |
0.05 |
| chr1_183134527_183134678 | 0.17 |
LAMC2 |
laminin, gamma 2 |
20771 |
0.19 |
| chr6_5671739_5671953 | 0.17 |
RP1-256G22.2 |
|
23659 |
0.27 |
| chr1_110576828_110577139 | 0.17 |
STRIP1 |
striatin interacting protein 1 |
255 |
0.89 |
| chr5_64775674_64775852 | 0.17 |
ADAMTS6 |
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
1941 |
0.4 |
| chr12_116893318_116893532 | 0.17 |
ENSG00000264037 |
. |
27302 |
0.23 |
| chr7_36195246_36195841 | 0.17 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
2666 |
0.32 |
| chr2_71694242_71694483 | 0.17 |
DYSF |
dysferlin |
530 |
0.84 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.3 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.2 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.3 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 0.1 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0070100 | Roundabout signaling pathway(GO:0035385) regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.3 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:1903312 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0043302 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.0 | GO:0033084 | regulation of immature T cell proliferation in thymus(GO:0033084) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.3 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0048048 | embryonic eye morphogenesis(GO:0048048) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.2 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.0 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.0 | GO:2000644 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0032610 | interleukin-1 alpha production(GO:0032610) |
| 0.0 | 0.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.0 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.0 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.0 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.0 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0043558 | regulation of translational initiation in response to stress(GO:0043558) |
| 0.0 | 0.1 | GO:0046132 | pyrimidine ribonucleoside biosynthetic process(GO:0046132) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.2 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0061043 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine development(GO:0060998) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.0 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.3 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |