Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
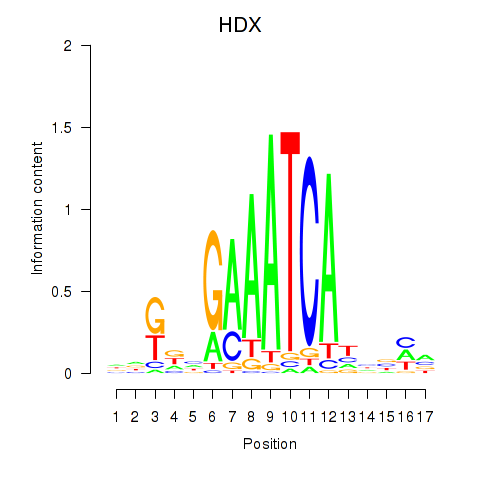
Results for HDX
Z-value: 0.83
Transcription factors associated with HDX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HDX
|
ENSG00000165259.9 | highly divergent homeobox |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_83756622_83756797 | HDX | 690 | 0.482330 | 0.51 | 1.6e-01 | Click! |
| chrX_83838309_83838460 | HDX | 80897 | 0.105276 | -0.40 | 2.9e-01 | Click! |
| chrX_83619155_83619306 | HDX | 138169 | 0.047244 | 0.39 | 3.1e-01 | Click! |
| chrX_83756396_83756547 | HDX | 928 | 0.445126 | -0.28 | 4.7e-01 | Click! |
| chrX_83756867_83757239 | HDX | 346 | 0.548016 | 0.10 | 8.0e-01 | Click! |
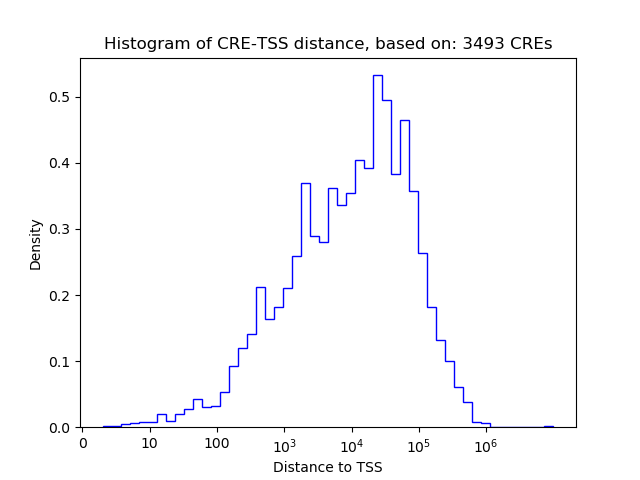
Activity of the HDX motif across conditions
Conditions sorted by the z-value of the HDX motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
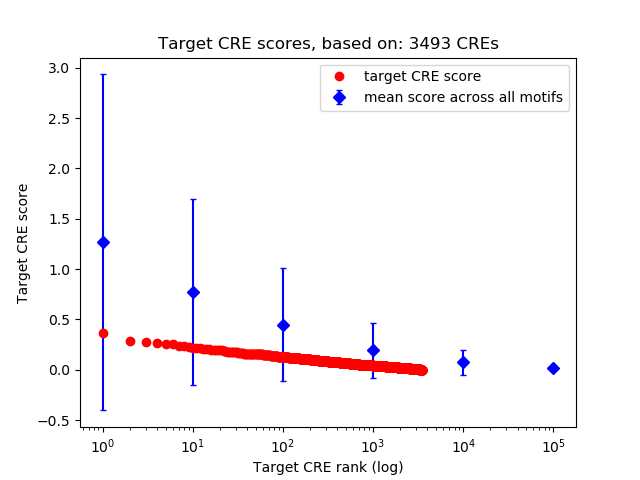
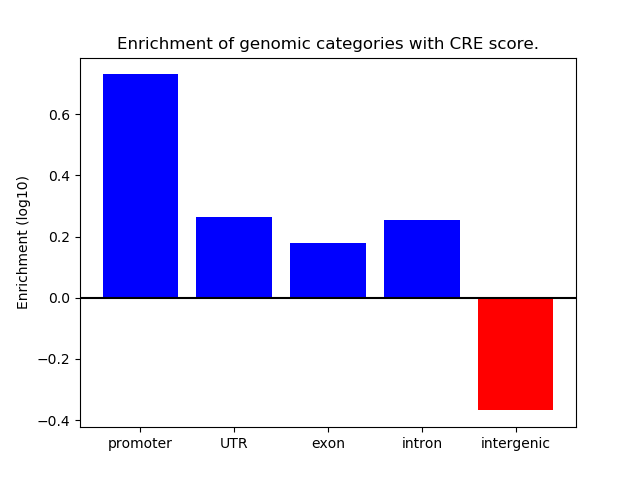
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_77380232_77380598 | 0.36 |
CTDP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
59386 |
0.13 |
| chr10_121579974_121580125 | 0.29 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
1821 |
0.39 |
| chr1_97004381_97004538 | 0.28 |
ENSG00000241992 |
. |
44306 |
0.2 |
| chr4_160169630_160170007 | 0.27 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
18215 |
0.23 |
| chr2_144339822_144340188 | 0.25 |
RP11-570L15.2 |
|
10331 |
0.21 |
| chr21_17951850_17952023 | 0.25 |
ENSG00000207863 |
. |
10621 |
0.23 |
| chr8_63998343_63998943 | 0.24 |
TTPA |
tocopherol (alpha) transfer protein |
31 |
0.98 |
| chr7_42377692_42377843 | 0.23 |
GLI3 |
GLI family zinc finger 3 |
101109 |
0.09 |
| chr4_154178869_154179109 | 0.22 |
TRIM2 |
tripartite motif containing 2 |
427 |
0.82 |
| chr17_59235574_59235725 | 0.22 |
RP11-136H19.1 |
|
22324 |
0.2 |
| chr5_107634116_107634267 | 0.22 |
FBXL17 |
F-box and leucine-rich repeat protein 17 |
69464 |
0.14 |
| chr18_34273309_34273594 | 0.21 |
FHOD3 |
formin homology 2 domain containing 3 |
25069 |
0.22 |
| chr19_7581346_7582140 | 0.21 |
ZNF358 |
zinc finger protein 358 |
739 |
0.45 |
| chr10_54326536_54326691 | 0.21 |
RP11-556E13.1 |
|
5748 |
0.34 |
| chr12_62529544_62529695 | 0.20 |
FAM19A2 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
198 |
0.95 |
| chr2_155312032_155312187 | 0.20 |
AC009227.2 |
|
1841 |
0.42 |
| chr8_89338531_89338740 | 0.20 |
RP11-586K2.1 |
|
430 |
0.76 |
| chr8_116675821_116675972 | 0.20 |
TRPS1 |
trichorhinophalangeal syndrome I |
1991 |
0.5 |
| chr3_21686525_21686676 | 0.20 |
ZNF385D-AS1 |
ZNF385D antisense RNA 1 |
102292 |
0.08 |
| chrY_7229387_7229538 | 0.19 |
ENSG00000252155 |
. |
17251 |
0.18 |
| chr17_80970638_80970845 | 0.19 |
RP11-1197K16.2 |
|
31783 |
0.15 |
| chr12_78334108_78334310 | 0.19 |
NAV3 |
neuron navigator 3 |
25847 |
0.27 |
| chr10_78733590_78733741 | 0.18 |
RP11-443A13.5 |
|
34799 |
0.21 |
| chr2_72438706_72438857 | 0.18 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
63614 |
0.16 |
| chr1_156098075_156098411 | 0.18 |
LMNA |
lamin A/C |
1649 |
0.24 |
| chr5_103680813_103681290 | 0.18 |
ENSG00000239808 |
. |
84524 |
0.11 |
| chr13_74899197_74899360 | 0.18 |
ENSG00000206617 |
. |
35927 |
0.21 |
| chrX_45634159_45634477 | 0.18 |
ENSG00000207725 |
. |
27788 |
0.21 |
| chr6_13463704_13463855 | 0.18 |
AL583828.1 |
|
22636 |
0.15 |
| chr15_100348989_100349200 | 0.18 |
CTD-2054N24.2 |
Uncharacterized protein |
901 |
0.5 |
| chr2_164592268_164592669 | 0.17 |
FIGN |
fidgetin |
49 |
0.99 |
| chr1_8194377_8194540 | 0.17 |
ENSG00000200975 |
. |
72199 |
0.1 |
| chr9_20873253_20873668 | 0.17 |
FOCAD |
focadhesin |
53167 |
0.14 |
| chr16_64944591_64944811 | 0.17 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
148880 |
0.05 |
| chr9_130614412_130614666 | 0.17 |
ENG |
endoglin |
2376 |
0.13 |
| chr12_88811798_88812120 | 0.16 |
ENSG00000199245 |
. |
12260 |
0.3 |
| chr2_8818889_8820001 | 0.16 |
AC011747.7 |
|
259 |
0.7 |
| chr12_15874929_15875080 | 0.16 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
6195 |
0.28 |
| chr14_64887783_64887949 | 0.16 |
CTD-2555O16.4 |
|
21090 |
0.12 |
| chr1_107805441_107805592 | 0.16 |
NTNG1 |
netrin G1 |
114300 |
0.07 |
| chr13_93105495_93105646 | 0.16 |
GPC5-IT1 |
GPC5 intronic transcript 1 (non-protein coding) |
31289 |
0.2 |
| chr6_12761420_12761571 | 0.16 |
ENSG00000223291 |
. |
7588 |
0.25 |
| chr14_27067998_27068288 | 0.16 |
NOVA1-AS1 |
NOVA1 antisense RNA 1 (head to head) |
525 |
0.68 |
| chr1_178995460_178996425 | 0.16 |
FAM20B |
family with sequence similarity 20, member B |
911 |
0.66 |
| chr10_133775388_133775627 | 0.16 |
BNIP3 |
BCL2/adenovirus E1B 19kDa interacting protein 3 |
19920 |
0.21 |
| chr2_223908392_223908543 | 0.16 |
KCNE4 |
potassium voltage-gated channel, Isk-related family, member 4 |
8065 |
0.28 |
| chr9_16628546_16628697 | 0.16 |
RP11-62F24.1 |
|
2947 |
0.36 |
| chr10_25008347_25008498 | 0.16 |
ARHGAP21 |
Rho GTPase activating protein 21 |
2373 |
0.41 |
| chr1_232803140_232803291 | 0.16 |
ENSG00000238382 |
. |
32735 |
0.22 |
| chr6_156979574_156979742 | 0.16 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
119405 |
0.06 |
| chr1_98515040_98515403 | 0.16 |
ENSG00000225206 |
. |
3494 |
0.38 |
| chr18_22928819_22929051 | 0.15 |
ZNF521 |
zinc finger protein 521 |
2222 |
0.42 |
| chr4_19731588_19731739 | 0.15 |
SLIT2 |
slit homolog 2 (Drosophila) |
523220 |
0.0 |
| chr8_24366846_24366997 | 0.15 |
RP11-561E1.1 |
|
5180 |
0.24 |
| chr5_140797768_140798225 | 0.15 |
PCDHGB7 |
protocadherin gamma subfamily B, 7 |
569 |
0.45 |
| chr7_55167054_55167326 | 0.15 |
EGFR |
epidermal growth factor receptor |
10226 |
0.29 |
| chr18_55447994_55448240 | 0.15 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
22210 |
0.18 |
| chr18_77378799_77378950 | 0.15 |
CTDP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
60927 |
0.13 |
| chr4_162286727_162286878 | 0.15 |
RP11-234O6.2 |
|
13303 |
0.32 |
| chr16_60157481_60157632 | 0.15 |
ENSG00000265167 |
. |
306081 |
0.01 |
| chr9_97666762_97667035 | 0.15 |
RP11-49O14.2 |
|
4187 |
0.23 |
| chr14_71372031_71372182 | 0.15 |
PCNX |
pecanex homolog (Drosophila) |
2016 |
0.46 |
| chr3_119219012_119219163 | 0.15 |
TIMMDC1 |
translocase of inner mitochondrial membrane domain containing 1 |
1659 |
0.33 |
| chr2_11076773_11076924 | 0.15 |
KCNF1 |
potassium voltage-gated channel, subfamily F, member 1 |
24785 |
0.2 |
| chr2_106912824_106912975 | 0.15 |
PLGLA |
plasminogen-like A (pseudogene) |
85663 |
0.09 |
| chr2_200025621_200025772 | 0.15 |
ENSG00000238698 |
. |
30086 |
0.25 |
| chr1_90018677_90018828 | 0.15 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
15048 |
0.22 |
| chr12_91430839_91430990 | 0.15 |
KERA |
keratocan |
20846 |
0.2 |
| chr10_15126962_15127123 | 0.14 |
ACBD7 |
acyl-CoA binding domain containing 7 |
3733 |
0.19 |
| chr1_119524620_119524878 | 0.14 |
TBX15 |
T-box 15 |
5679 |
0.29 |
| chr17_13265618_13265967 | 0.14 |
ENSG00000266115 |
. |
137853 |
0.05 |
| chr12_91504528_91504679 | 0.14 |
LUM |
lumican |
1005 |
0.61 |
| chr4_41157747_41157945 | 0.14 |
ENSG00000207198 |
. |
41887 |
0.14 |
| chr18_51886872_51887023 | 0.14 |
C18orf54 |
chromosome 18 open reading frame 54 |
1992 |
0.33 |
| chr1_200212018_200212550 | 0.14 |
ENSG00000221403 |
. |
98322 |
0.08 |
| chr3_178015611_178015762 | 0.14 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
24966 |
0.26 |
| chr2_180214031_180214182 | 0.14 |
SESTD1 |
SEC14 and spectrin domains 1 |
84589 |
0.1 |
| chr6_35182117_35182910 | 0.14 |
SCUBE3 |
signal peptide, CUB domain, EGF-like 3 |
317 |
0.9 |
| chr12_132316032_132316383 | 0.14 |
MMP17 |
matrix metallopeptidase 17 (membrane-inserted) |
3122 |
0.22 |
| chr4_17325894_17326097 | 0.14 |
ENSG00000206780 |
. |
3490 |
0.36 |
| chr13_45145551_45145847 | 0.14 |
TSC22D1 |
TSC22 domain family, member 1 |
4693 |
0.31 |
| chr16_86619409_86619937 | 0.14 |
FOXL1 |
forkhead box L1 |
7558 |
0.18 |
| chr4_148024489_148024820 | 0.13 |
TTC29 |
tetratricopeptide repeat domain 29 |
157620 |
0.04 |
| chr2_101359270_101359474 | 0.13 |
NPAS2 |
neuronal PAS domain protein 2 |
77242 |
0.09 |
| chr3_88191761_88191912 | 0.13 |
ZNF654 |
zinc finger protein 654 |
3582 |
0.23 |
| chr6_56818301_56819485 | 0.13 |
DST |
dystonin |
492 |
0.63 |
| chr17_17862779_17863058 | 0.13 |
TOM1L2 |
target of myb1-like 2 (chicken) |
12793 |
0.15 |
| chr20_52273531_52273682 | 0.13 |
ENSG00000238468 |
. |
11691 |
0.23 |
| chr9_89336989_89337198 | 0.13 |
GAS1 |
growth arrest-specific 1 |
225011 |
0.02 |
| chr17_15209661_15209932 | 0.13 |
ENSG00000200437 |
. |
4866 |
0.18 |
| chr11_114398277_114398428 | 0.13 |
ENSG00000238724 |
. |
501 |
0.77 |
| chr6_157297907_157298141 | 0.13 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
75517 |
0.12 |
| chr6_155783008_155783159 | 0.13 |
NOX3 |
NADPH oxidase 3 |
6046 |
0.32 |
| chr2_212025965_212026310 | 0.13 |
ENSG00000199585 |
. |
374241 |
0.01 |
| chr5_980561_980809 | 0.13 |
RP11-661C8.3 |
Uncharacterized protein |
1205 |
0.39 |
| chr19_31153950_31154101 | 0.13 |
ENSG00000223148 |
. |
48898 |
0.19 |
| chrX_131467583_131467734 | 0.13 |
MBNL3 |
muscleblind-like splicing regulator 3 |
57330 |
0.15 |
| chr17_63693222_63693493 | 0.13 |
CEP112 |
centrosomal protein 112kDa |
53485 |
0.17 |
| chr8_126010804_126011981 | 0.13 |
SQLE |
squalene epoxidase |
606 |
0.56 |
| chr3_159978482_159978633 | 0.13 |
RP11-431I8.1 |
|
34287 |
0.13 |
| chr2_26401607_26402234 | 0.13 |
GAREML |
GRB2 associated, regulator of MAPK1-like |
1665 |
0.38 |
| chr8_20080573_20080724 | 0.13 |
ENSG00000222267 |
. |
6642 |
0.19 |
| chr2_17484452_17484603 | 0.13 |
ENSG00000222842 |
. |
180120 |
0.03 |
| chr9_91138470_91138621 | 0.13 |
NXNL2 |
nucleoredoxin-like 2 |
11471 |
0.29 |
| chr1_84629927_84630112 | 0.13 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
13 |
0.99 |
| chr17_37099094_37099245 | 0.13 |
RP1-56K13.3 |
|
21147 |
0.11 |
| chr8_120648373_120648673 | 0.13 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
2497 |
0.38 |
| chr14_33178041_33178192 | 0.13 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
26941 |
0.27 |
| chr2_20098121_20098272 | 0.12 |
TTC32 |
tetratricopeptide repeat domain 32 |
3206 |
0.31 |
| chr8_117529471_117529622 | 0.12 |
ENSG00000264815 |
. |
229958 |
0.02 |
| chr12_64580629_64580780 | 0.12 |
ENSG00000212298 |
. |
17869 |
0.15 |
| chr6_148886558_148886709 | 0.12 |
ENSG00000223322 |
. |
41257 |
0.21 |
| chrX_13680096_13680380 | 0.12 |
TCEANC |
transcription elongation factor A (SII) N-terminal and central domain containing |
6824 |
0.2 |
| chr6_17925089_17925240 | 0.12 |
KIF13A |
kinesin family member 13A |
62530 |
0.13 |
| chr13_73356894_73357293 | 0.12 |
DIS3 |
DIS3 mitotic control homolog (S. cerevisiae) |
859 |
0.4 |
| chr5_64775674_64775852 | 0.12 |
ADAMTS6 |
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
1941 |
0.4 |
| chr4_175056009_175056160 | 0.12 |
FBXO8 |
F-box protein 8 |
148730 |
0.04 |
| chr13_36298655_36298806 | 0.12 |
DCLK1 |
doublecortin-like kinase 1 |
131049 |
0.05 |
| chr18_77379773_77379948 | 0.12 |
CTDP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
59941 |
0.13 |
| chr2_9778930_9779081 | 0.12 |
YWHAQ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
7862 |
0.21 |
| chr8_99410632_99410929 | 0.12 |
KCNS2 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
28470 |
0.18 |
| chr12_21981834_21982050 | 0.12 |
RP11-729I10.2 |
|
1798 |
0.39 |
| chr13_31482253_31482459 | 0.12 |
MEDAG |
mesenteric estrogen-dependent adipogenesis |
1513 |
0.41 |
| chr10_36783162_36783313 | 0.12 |
NAMPTL |
nicotinamide phosphoribosyltransferase-like |
29086 |
0.22 |
| chr7_15596833_15597046 | 0.12 |
AGMO |
alkylglycerol monooxygenase |
4701 |
0.34 |
| chr2_204679921_204680089 | 0.12 |
ENSG00000206970 |
. |
33245 |
0.17 |
| chr16_67203881_67204335 | 0.12 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
51 |
0.91 |
| chr1_99731775_99731926 | 0.12 |
LPPR4 |
Lipid phosphate phosphatase-related protein type 4 |
2002 |
0.49 |
| chr11_107461958_107462688 | 0.12 |
AP000889.3 |
HCG2032453; Uncharacterized protein; cDNA FLJ25337 fis, clone TST00714 |
148 |
0.74 |
| chr8_72777208_72777359 | 0.12 |
MSC |
musculin |
20580 |
0.17 |
| chr2_75867971_75868176 | 0.12 |
ENSG00000239018 |
. |
5806 |
0.13 |
| chr8_117883179_117883330 | 0.12 |
RAD21 |
RAD21 homolog (S. pombe) |
3345 |
0.19 |
| chr20_61656853_61657255 | 0.12 |
BHLHE23 |
basic helix-loop-helix family, member e23 |
18667 |
0.17 |
| chr17_79317268_79317735 | 0.12 |
TMEM105 |
transmembrane protein 105 |
13027 |
0.13 |
| chr13_47470589_47471243 | 0.12 |
HTR2A |
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
253 |
0.95 |
| chr10_73946584_73946735 | 0.12 |
ENSG00000200294 |
. |
6043 |
0.17 |
| chr7_33111925_33112076 | 0.12 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
9591 |
0.15 |
| chr2_119603311_119603908 | 0.12 |
EN1 |
engrailed homeobox 1 |
1645 |
0.51 |
| chr4_152146952_152147499 | 0.11 |
SH3D19 |
SH3 domain containing 19 |
7 |
0.98 |
| chr2_39719567_39719774 | 0.11 |
AC007246.3 |
|
21617 |
0.22 |
| chr18_3625070_3625338 | 0.11 |
DLGAP1-AS2 |
DLGAP1 antisense RNA 2 |
21468 |
0.14 |
| chr13_36274861_36275012 | 0.11 |
NBEA |
neurobeachin |
107793 |
0.07 |
| chr12_95177906_95178057 | 0.11 |
ENSG00000208038 |
. |
50193 |
0.17 |
| chr5_158559403_158559554 | 0.11 |
EBF1 |
early B-cell factor 1 |
32709 |
0.18 |
| chr12_113709895_113710081 | 0.11 |
TPCN1 |
two pore segment channel 1 |
4735 |
0.18 |
| chr8_117456913_117457110 | 0.11 |
ENSG00000264815 |
. |
157423 |
0.04 |
| chr3_174044116_174044267 | 0.11 |
NAALADL2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
114586 |
0.07 |
| chr4_16198053_16198212 | 0.11 |
TAPT1 |
transmembrane anterior posterior transformation 1 |
5025 |
0.22 |
| chr3_193851589_193851867 | 0.11 |
HES1 |
hes family bHLH transcription factor 1 |
2206 |
0.28 |
| chr3_28957653_28958300 | 0.11 |
ENSG00000238470 |
. |
94757 |
0.09 |
| chr9_18386232_18386424 | 0.11 |
ADAMTSL1 |
ADAMTS-like 1 |
87564 |
0.1 |
| chr6_17868077_17868228 | 0.11 |
KIF13A |
kinesin family member 13A |
50721 |
0.16 |
| chr18_18944422_18944573 | 0.11 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
943 |
0.63 |
| chr15_101632383_101632547 | 0.11 |
RP11-505E24.2 |
|
6194 |
0.25 |
| chr13_75940906_75941057 | 0.11 |
TBC1D4-AS1 |
TBC1D4 antisense RNA 1 |
10449 |
0.24 |
| chr4_53719384_53719535 | 0.11 |
RASL11B |
RAS-like, family 11, member B |
8998 |
0.22 |
| chr6_138002128_138002279 | 0.11 |
ENSG00000216097 |
. |
35882 |
0.2 |
| chr2_201331503_201331739 | 0.11 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
26249 |
0.16 |
| chr17_47078770_47079113 | 0.11 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
3833 |
0.12 |
| chr1_67002413_67002591 | 0.11 |
SGIP1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
2537 |
0.38 |
| chr8_39965880_39966091 | 0.11 |
C8orf4 |
chromosome 8 open reading frame 4 |
45004 |
0.18 |
| chr1_162596970_162597121 | 0.11 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
4118 |
0.23 |
| chr7_92327286_92327437 | 0.11 |
ENSG00000206763 |
. |
3767 |
0.31 |
| chr14_52348217_52348368 | 0.11 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
3982 |
0.24 |
| chr4_138452050_138452321 | 0.11 |
PCDH18 |
protocadherin 18 |
1380 |
0.62 |
| chr11_132614961_132615112 | 0.11 |
OPCML-IT2 |
OPCML intronic transcript 2 (non-protein coding) |
114936 |
0.07 |
| chr5_152825234_152825385 | 0.11 |
GRIA1 |
glutamate receptor, ionotropic, AMPA 1 |
44797 |
0.21 |
| chr8_138161471_138161805 | 0.11 |
ENSG00000206678 |
. |
44043 |
0.22 |
| chr16_54465247_54465407 | 0.11 |
ENSG00000264079 |
. |
117981 |
0.06 |
| chrX_103554920_103555123 | 0.11 |
ESX1 |
ESX homeobox 1 |
55407 |
0.15 |
| chr2_11508011_11508277 | 0.11 |
ROCK2 |
Rho-associated, coiled-coil containing protein kinase 2 |
23433 |
0.2 |
| chr10_63595192_63595343 | 0.11 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
65792 |
0.13 |
| chr14_55239224_55239463 | 0.11 |
SAMD4A |
sterile alpha motif domain containing 4A |
17792 |
0.24 |
| chr7_139578044_139578195 | 0.11 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
49009 |
0.16 |
| chr15_48936207_48936433 | 0.11 |
FBN1 |
fibrillin 1 |
1598 |
0.51 |
| chr3_114119404_114119777 | 0.11 |
ZBTB20-AS1 |
ZBTB20 antisense RNA 1 |
13074 |
0.21 |
| chr2_27434850_27435940 | 0.11 |
SLC5A6 |
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
5 |
0.66 |
| chr12_91721216_91721367 | 0.11 |
DCN |
decorin |
144391 |
0.05 |
| chr3_77562048_77562199 | 0.11 |
ROBO2 |
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
75809 |
0.13 |
| chr20_51658849_51659000 | 0.11 |
ENSG00000252629 |
. |
20232 |
0.22 |
| chr6_157956725_157956898 | 0.11 |
ENSG00000266617 |
. |
6647 |
0.28 |
| chr4_87812935_87814058 | 0.11 |
C4orf36 |
chromosome 4 open reading frame 36 |
52 |
0.98 |
| chr6_24283092_24283303 | 0.10 |
KAAG1 |
kidney associated antigen 1 |
73934 |
0.09 |
| chr15_36145502_36145653 | 0.10 |
ENSG00000265098 |
. |
73480 |
0.13 |
| chr7_27680791_27681081 | 0.10 |
HIBADH |
3-hydroxyisobutyrate dehydrogenase |
6439 |
0.28 |
| chr4_169553568_169553728 | 0.10 |
PALLD |
palladin, cytoskeletal associated protein |
880 |
0.63 |
| chr2_1658470_1658621 | 0.10 |
AC144450.1 |
|
34660 |
0.19 |
| chr8_38316900_38317069 | 0.10 |
FGFR1 |
fibroblast growth factor receptor 1 |
1963 |
0.28 |
| chr10_3495258_3495563 | 0.10 |
PITRM1 |
pitrilysin metallopeptidase 1 |
280407 |
0.01 |
| chr4_169060384_169060535 | 0.10 |
RP11-310I9.1 |
|
25977 |
0.21 |
| chr9_34654990_34655141 | 0.10 |
IL11RA |
interleukin 11 receptor, alpha |
16 |
0.94 |
| chr12_70132575_70133879 | 0.10 |
RAB3IP |
RAB3A interacting protein |
47 |
0.95 |
| chr7_82052951_82053102 | 0.10 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
20005 |
0.29 |
| chr7_111079006_111079157 | 0.10 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
46110 |
0.2 |
| chr10_711808_711959 | 0.10 |
RP11-809C18.5 |
|
2385 |
0.24 |
| chr7_137026886_137027037 | 0.10 |
PTN |
pleiotrophin |
1559 |
0.52 |
| chr7_93518213_93518642 | 0.10 |
TFPI2 |
tissue factor pathway inhibitor 2 |
1055 |
0.43 |
| chr8_42013507_42013690 | 0.10 |
AP3M2 |
adaptor-related protein complex 3, mu 2 subunit |
1464 |
0.36 |
| chr7_24517307_24517482 | 0.10 |
ENSG00000206877 |
. |
59557 |
0.14 |
| chr1_234879912_234880063 | 0.10 |
ENSG00000201638 |
. |
93733 |
0.08 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.0 | GO:2000380 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.2 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.2 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.0 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) neural crest cell fate commitment(GO:0014034) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.1 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.0 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0003207 | cardiac chamber formation(GO:0003207) cardiac ventricle formation(GO:0003211) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0046970 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.0 | REACTOME PI 3K CASCADE | Genes involved in PI-3K cascade |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |