Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
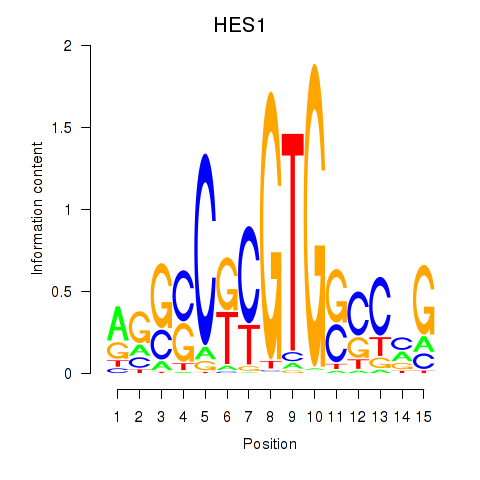
Results for HES1
Z-value: 0.69
Transcription factors associated with HES1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HES1
|
ENSG00000114315.3 | hes family bHLH transcription factor 1 |
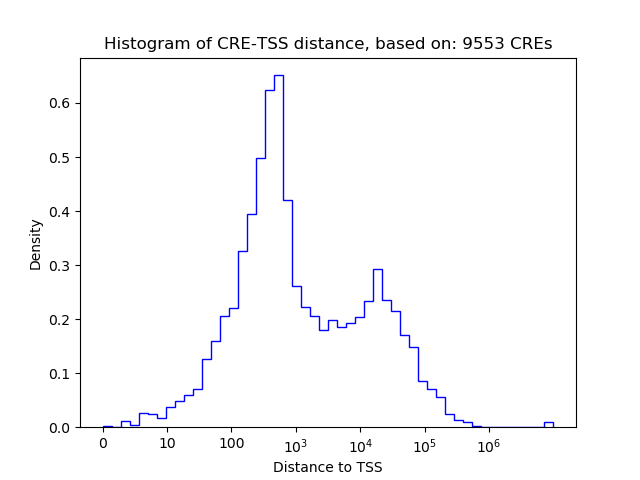
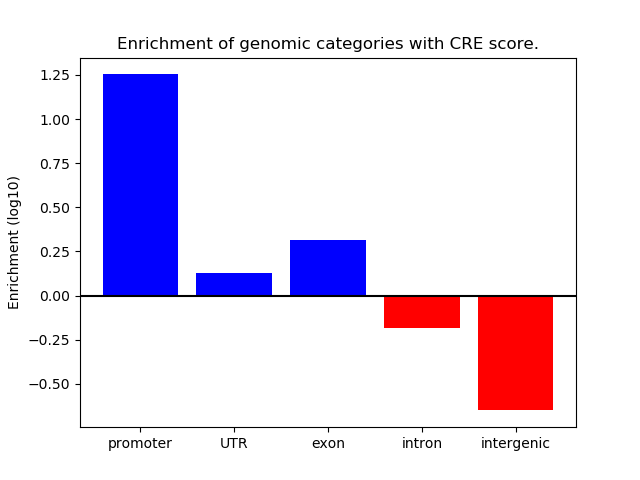
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_193851589_193851867 | HES1 | 2206 | 0.279822 | 0.61 | 8.1e-02 | Click! |
| chr3_193855921_193856072 | HES1 | 2062 | 0.312901 | 0.47 | 2.0e-01 | Click! |
| chr3_193852202_193852818 | HES1 | 1424 | 0.403707 | -0.42 | 2.6e-01 | Click! |
| chr3_193857154_193857355 | HES1 | 3320 | 0.237261 | 0.39 | 3.0e-01 | Click! |
| chr3_193857797_193857985 | HES1 | 3957 | 0.220496 | 0.37 | 3.2e-01 | Click! |
Activity of the HES1 motif across conditions
Conditions sorted by the z-value of the HES1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
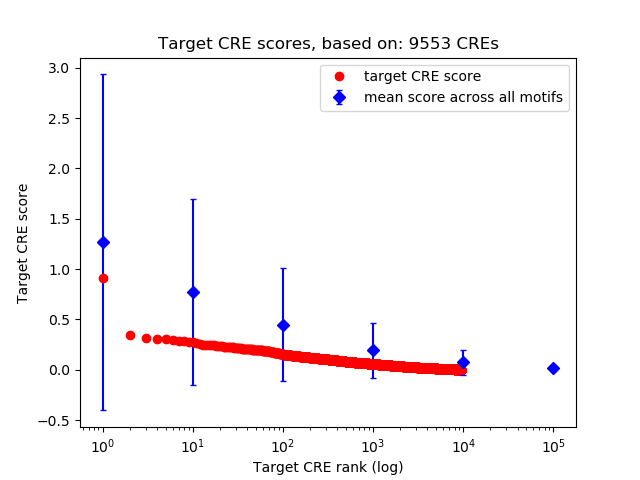
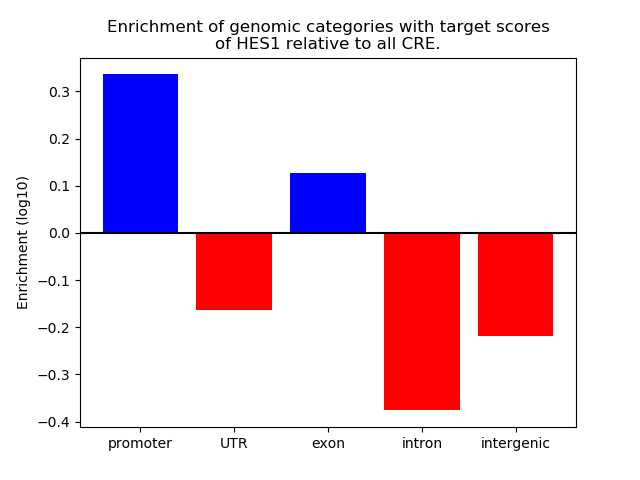
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_112031428_112032221 | 0.92 |
AC004112.4 |
|
17854 |
0.19 |
| chr7_5466302_5466453 | 0.35 |
TNRC18 |
trinucleotide repeat containing 18 |
1332 |
0.35 |
| chr13_29068702_29069490 | 0.32 |
FLT1 |
fms-related tyrosine kinase 1 |
136 |
0.98 |
| chr9_132382165_132382732 | 0.31 |
C9orf50 |
chromosome 9 open reading frame 50 |
607 |
0.62 |
| chr1_66258526_66259233 | 0.30 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
15 |
0.99 |
| chr17_79318428_79319403 | 0.29 |
TMEM105 |
transmembrane protein 105 |
14441 |
0.13 |
| chr22_41633466_41634617 | 0.29 |
CHADL |
chondroadherin-like |
1584 |
0.26 |
| chr8_69242814_69243988 | 0.28 |
C8orf34 |
chromosome 8 open reading frame 34 |
56 |
0.82 |
| chr2_144694678_144695023 | 0.27 |
AC016910.1 |
|
210 |
0.96 |
| chr16_55542879_55544136 | 0.27 |
LPCAT2 |
lysophosphatidylcholine acyltransferase 2 |
597 |
0.77 |
| chr14_37667189_37667862 | 0.26 |
MIPOL1 |
mirror-image polydactyly 1 |
232 |
0.92 |
| chr7_5466974_5467725 | 0.25 |
TNRC18 |
trinucleotide repeat containing 18 |
2304 |
0.22 |
| chr7_15725092_15725754 | 0.25 |
MEOX2 |
mesenchyme homeobox 2 |
1014 |
0.63 |
| chrX_12156590_12157176 | 0.25 |
FRMPD4 |
FERM and PDZ domain containing 4 |
298 |
0.9 |
| chr3_181412989_181413321 | 0.25 |
SOX2-OT |
SOX2 overlapping transcript (non-protein coding) |
4126 |
0.28 |
| chr22_46659115_46659367 | 0.25 |
PKDREJ |
polycystin (PKD) family receptor for egg jelly |
22 |
0.97 |
| chr10_11784750_11785238 | 0.25 |
ECHDC3 |
enoyl CoA hydratase domain containing 3 |
549 |
0.83 |
| chr18_31020374_31020722 | 0.24 |
CCDC178 |
coiled-coil domain containing 178 |
109 |
0.98 |
| chr4_151500979_151501427 | 0.24 |
RP11-1336O20.2 |
|
962 |
0.54 |
| chr21_40032373_40032758 | 0.23 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
26 |
0.99 |
| chr1_14924992_14926197 | 0.23 |
KAZN |
kazrin, periplakin interacting protein |
381 |
0.93 |
| chr20_11871763_11872529 | 0.23 |
BTBD3 |
BTB (POZ) domain containing 3 |
171 |
0.97 |
| chr8_144512945_144513817 | 0.23 |
MAFA |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A |
805 |
0.48 |
| chr3_8809480_8810320 | 0.23 |
OXTR |
oxytocin receptor |
463 |
0.76 |
| chr7_193046_193319 | 0.23 |
FAM20C |
family with sequence similarity 20, member C |
213 |
0.89 |
| chr3_196729783_196730213 | 0.23 |
MFI2-AS1 |
MFI2 antisense RNA 1 |
5 |
0.97 |
| chr14_89883615_89884477 | 0.22 |
FOXN3-AS1 |
FOXN3 antisense RNA 1 |
348 |
0.65 |
| chr3_98451018_98451517 | 0.22 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
27 |
0.56 |
| chr8_93114852_93115389 | 0.22 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
334 |
0.94 |
| chr17_79448132_79448599 | 0.22 |
RP11-1055B8.8 |
|
19402 |
0.09 |
| chr16_3095827_3096553 | 0.22 |
MMP25 |
matrix metallopeptidase 25 |
492 |
0.51 |
| chr5_63461313_63462499 | 0.22 |
RNF180 |
ring finger protein 180 |
83 |
0.99 |
| chr2_105458782_105459564 | 0.21 |
LINC01158 |
long intergenic non-protein coding RNA 1158 |
8738 |
0.14 |
| chr4_57975609_57976167 | 0.21 |
IGFBP7-AS1 |
IGFBP7 antisense RNA 1 |
40 |
0.93 |
| chr11_74442538_74442948 | 0.21 |
CHRDL2 |
chordin-like 2 |
313 |
0.84 |
| chr8_25042452_25043634 | 0.21 |
DOCK5 |
dedicator of cytokinesis 5 |
663 |
0.75 |
| chr10_126106510_126107495 | 0.21 |
OAT |
ornithine aminotransferase |
503 |
0.8 |
| chr20_11871067_11871503 | 0.21 |
BTBD3 |
BTB (POZ) domain containing 3 |
86 |
0.98 |
| chr8_38614830_38615236 | 0.21 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
182 |
0.95 |
| chr2_68545863_68546629 | 0.20 |
CNRIP1 |
cannabinoid receptor interacting protein 1 |
286 |
0.89 |
| chr22_22011512_22012681 | 0.20 |
KB-1440D3.13 |
|
4127 |
0.1 |
| chr7_23513498_23513922 | 0.20 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
3624 |
0.21 |
| chr3_157260663_157261197 | 0.20 |
C3orf55 |
chromosome 3 open reading frame 55 |
105 |
0.98 |
| chrX_38079311_38080259 | 0.20 |
SRPX |
sushi-repeat containing protein, X-linked |
367 |
0.75 |
| chr7_143582018_143582424 | 0.20 |
FAM115A |
family with sequence similarity 115, member A |
242 |
0.92 |
| chr12_49627444_49627663 | 0.20 |
TUBA1C |
tubulin, alpha 1c |
5844 |
0.13 |
| chr5_131862398_131862590 | 0.20 |
IL5 |
interleukin 5 (colony-stimulating factor, eosinophil) |
16720 |
0.13 |
| chr17_76164337_76165654 | 0.20 |
SYNGR2 |
synaptogyrin 2 |
244 |
0.87 |
| chr18_21851258_21851803 | 0.20 |
OSBPL1A |
oxysterol binding protein-like 1A |
666 |
0.6 |
| chr12_3308931_3310293 | 0.20 |
TSPAN9 |
tetraspanin 9 |
729 |
0.75 |
| chr3_192126971_192127690 | 0.20 |
FGF12 |
fibroblast growth factor 12 |
492 |
0.87 |
| chr17_59478776_59479020 | 0.20 |
RP11-332H18.5 |
|
1294 |
0.27 |
| chr9_132382776_132383013 | 0.20 |
C9orf50 |
chromosome 9 open reading frame 50 |
161 |
0.92 |
| chr2_121493751_121494304 | 0.20 |
GLI2 |
GLI family zinc finger 2 |
204 |
0.97 |
| chr1_205648469_205649627 | 0.19 |
SLC45A3 |
solute carrier family 45, member 3 |
539 |
0.74 |
| chr3_24562978_24563646 | 0.19 |
ENSG00000265028 |
. |
386 |
0.87 |
| chr14_61104436_61104673 | 0.19 |
SIX1 |
SIX homeobox 1 |
11626 |
0.21 |
| chr20_6032553_6033205 | 0.19 |
LRRN4 |
leucine rich repeat neuronal 4 |
1816 |
0.35 |
| chr20_23617810_23618640 | 0.19 |
CST3 |
cystatin C |
357 |
0.87 |
| chr9_94183938_94184089 | 0.19 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
2131 |
0.44 |
| chr9_139715640_139716017 | 0.19 |
RABL6 |
RAB, member RAS oncogene family-like 6 |
47 |
0.93 |
| chr2_242497891_242499073 | 0.19 |
BOK-AS1 |
BOK antisense RNA 1 |
90 |
0.79 |
| chr13_37005266_37006411 | 0.19 |
CCNA1 |
cyclin A1 |
129 |
0.97 |
| chr11_119019412_119020504 | 0.19 |
ABCG4 |
ATP-binding cassette, sub-family G (WHITE), member 4 |
5 |
0.94 |
| chr7_156400256_156401034 | 0.19 |
ENSG00000182648 |
. |
2187 |
0.32 |
| chr11_62455448_62455813 | 0.19 |
LRRN4CL |
LRRN4 C-terminal like |
1741 |
0.12 |
| chr3_168864106_168864430 | 0.19 |
MECOM |
MDS1 and EVI1 complex locus |
59 |
0.99 |
| chr7_89949941_89950814 | 0.19 |
C7orf63 |
chromosome 7 open reading frame 63 |
13986 |
0.16 |
| chr20_49347136_49348386 | 0.18 |
PARD6B |
par-6 family cell polarity regulator beta |
320 |
0.89 |
| chr1_223743422_223744240 | 0.18 |
CAPN8 |
calpain 8 |
19451 |
0.25 |
| chr9_139742382_139743338 | 0.18 |
PHPT1 |
phosphohistidine phosphatase 1 |
316 |
0.71 |
| chr4_128543868_128544388 | 0.18 |
INTU |
inturned planar cell polarity protein |
298 |
0.94 |
| chr7_23511015_23511471 | 0.18 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
1157 |
0.46 |
| chr5_68788670_68789791 | 0.18 |
OCLN |
occludin |
612 |
0.62 |
| chr20_30457693_30458808 | 0.17 |
DUSP15 |
dual specificity phosphatase 15 |
125 |
0.67 |
| chr5_123987558_123987770 | 0.17 |
RP11-436H11.2 |
|
76860 |
0.08 |
| chr13_29106539_29107199 | 0.17 |
FLT1 |
fms-related tyrosine kinase 1 |
37604 |
0.21 |
| chr12_95043269_95044318 | 0.17 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
545 |
0.85 |
| chr17_79369714_79370212 | 0.17 |
RP11-1055B8.6 |
Uncharacterized protein |
688 |
0.54 |
| chr6_46702576_46703631 | 0.17 |
PLA2G7 |
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
24 |
0.97 |
| chr16_54317621_54317889 | 0.17 |
IRX3 |
iroquois homeobox 3 |
2012 |
0.4 |
| chr21_46962488_46963290 | 0.17 |
SLC19A1 |
solute carrier family 19 (folate transporter), member 1 |
504 |
0.83 |
| chr3_127540841_127541975 | 0.17 |
MGLL |
monoglyceride lipase |
214 |
0.96 |
| chr1_21836241_21836468 | 0.17 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
489 |
0.83 |
| chr5_76475983_76476746 | 0.17 |
PDE8B |
phosphodiesterase 8B |
29910 |
0.15 |
| chr19_13906290_13907522 | 0.16 |
ZSWIM4 |
zinc finger, SWIM-type containing 4 |
632 |
0.39 |
| chr15_66914300_66914605 | 0.16 |
RP11-321F6.1 |
HCG2003567; Uncharacterized protein |
39924 |
0.12 |
| chr19_45655667_45657201 | 0.16 |
NKPD1 |
NTPase, KAP family P-loop domain containing 1 |
594 |
0.58 |
| chr9_127539201_127540134 | 0.16 |
OLFML2A |
olfactomedin-like 2A |
117 |
0.95 |
| chr8_49782515_49783288 | 0.16 |
SNAI2 |
snail family zinc finger 2 |
51087 |
0.18 |
| chr2_133061942_133062584 | 0.16 |
ENSG00000221288 |
. |
47610 |
0.14 |
| chr17_78044749_78045392 | 0.16 |
CCDC40 |
coiled-coil domain containing 40 |
12346 |
0.15 |
| chr8_687260_687856 | 0.16 |
ERICH1-AS1 |
ERICH1 antisense RNA 1 |
93 |
0.97 |
| chr16_85783565_85784591 | 0.16 |
C16orf74 |
chromosome 16 open reading frame 74 |
479 |
0.7 |
| chr15_66994787_66995642 | 0.16 |
SMAD6 |
SMAD family member 6 |
540 |
0.83 |
| chr1_180881914_180882730 | 0.16 |
KIAA1614 |
KIAA1614 |
3 |
0.98 |
| chr6_35182117_35182910 | 0.16 |
SCUBE3 |
signal peptide, CUB domain, EGF-like 3 |
317 |
0.9 |
| chr1_215740105_215740888 | 0.16 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
239 |
0.96 |
| chr9_86152792_86153773 | 0.16 |
FRMD3 |
FERM domain containing 3 |
71 |
0.98 |
| chr14_102094993_102095193 | 0.15 |
DIO3 |
deiodinase, iodothyronine, type III |
67405 |
0.09 |
| chr4_682374_683342 | 0.15 |
MFSD7 |
major facilitator superfamily domain containing 7 |
74 |
0.94 |
| chr2_172944774_172945201 | 0.15 |
DLX1 |
distal-less homeobox 1 |
4481 |
0.24 |
| chr2_185463143_185463634 | 0.15 |
ZNF804A |
zinc finger protein 804A |
295 |
0.95 |
| chr10_44879737_44880262 | 0.15 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
492 |
0.84 |
| chr7_38624277_38624485 | 0.15 |
AMPH |
amphiphysin |
46639 |
0.16 |
| chr6_19691587_19692480 | 0.15 |
ENSG00000200957 |
. |
49273 |
0.18 |
| chrX_153199314_153200499 | 0.15 |
NAA10 |
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
505 |
0.6 |
| chr9_16063260_16063427 | 0.15 |
C9orf92 |
chromosome 9 open reading frame 92 |
152554 |
0.04 |
| chr20_32273512_32274295 | 0.15 |
E2F1 |
E2F transcription factor 1 |
307 |
0.81 |
| chr16_88450130_88450579 | 0.15 |
ZNF469 |
zinc finger protein 469 |
43525 |
0.15 |
| chr18_74322772_74323202 | 0.15 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
82113 |
0.09 |
| chr1_110693132_110693795 | 0.15 |
SLC6A17 |
solute carrier family 6 (neutral amino acid transporter), member 17 |
355 |
0.81 |
| chrX_102318388_102319159 | 0.15 |
BEX1 |
brain expressed, X-linked 1 |
395 |
0.83 |
| chr2_172379612_172380165 | 0.15 |
CYBRD1 |
cytochrome b reductase 1 |
299 |
0.93 |
| chr16_57513132_57513388 | 0.15 |
DOK4 |
docking protein 4 |
457 |
0.75 |
| chr19_51871753_51871904 | 0.15 |
CLDND2 |
claudin domain containing 2 |
405 |
0.61 |
| chr6_105388737_105389249 | 0.15 |
LIN28B |
lin-28 homolog B (C. elegans) |
15930 |
0.23 |
| chr12_51236674_51236895 | 0.15 |
TMPRSS12 |
transmembrane (C-terminal) protease, serine 12 |
68 |
0.95 |
| chr4_6927779_6927930 | 0.14 |
TBC1D14 |
TBC1 domain family, member 14 |
15879 |
0.16 |
| chr1_162531480_162532245 | 0.14 |
UAP1 |
UDP-N-acteylglucosamine pyrophosphorylase 1 |
539 |
0.55 |
| chr2_69968563_69969704 | 0.14 |
ANXA4 |
annexin A4 |
27 |
0.98 |
| chr8_116675050_116675578 | 0.14 |
TRPS1 |
trichorhinophalangeal syndrome I |
1409 |
0.6 |
| chr10_19777990_19778239 | 0.14 |
MALRD1 |
MAM and LDL receptor class A domain containing 1 |
91 |
0.98 |
| chr9_115774055_115774548 | 0.14 |
ZFP37 |
ZFP37 zinc finger protein |
44667 |
0.16 |
| chrX_53349448_53350467 | 0.14 |
IQSEC2 |
IQ motif and Sec7 domain 2 |
565 |
0.73 |
| chr10_71812550_71812868 | 0.14 |
H2AFY2 |
H2A histone family, member Y2 |
73 |
0.98 |
| chr15_47476270_47477144 | 0.14 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
409 |
0.87 |
| chr1_240161229_240161618 | 0.14 |
FMN2 |
formin 2 |
16225 |
0.27 |
| chr17_31255277_31255844 | 0.14 |
TMEM98 |
transmembrane protein 98 |
332 |
0.89 |
| chr3_196415381_196415617 | 0.14 |
ENSG00000201441 |
. |
20336 |
0.11 |
| chr3_158449888_158450546 | 0.14 |
RARRES1 |
retinoic acid receptor responder (tazarotene induced) 1 |
14 |
0.36 |
| chr11_89520365_89520516 | 0.14 |
TRIM64DP |
tripartite motif containing 64D, pseudogene |
10291 |
0.21 |
| chr11_109964287_109965206 | 0.14 |
ZC3H12C |
zinc finger CCCH-type containing 12C |
659 |
0.83 |
| chr2_236403404_236403896 | 0.14 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
899 |
0.65 |
| chr10_77168132_77169350 | 0.14 |
ENSG00000237149 |
. |
5929 |
0.2 |
| chr9_88713671_88714373 | 0.14 |
GOLM1 |
golgi membrane protein 1 |
451 |
0.9 |
| chr5_153990189_153990659 | 0.14 |
ENSG00000264760 |
. |
14792 |
0.21 |
| chr10_101088892_101089952 | 0.14 |
CNNM1 |
cyclin M1 |
266 |
0.95 |
| chr7_127672748_127672947 | 0.14 |
LRRC4 |
leucine rich repeat containing 4 |
687 |
0.76 |
| chr11_69453965_69454310 | 0.14 |
CCND1 |
cyclin D1 |
1718 |
0.41 |
| chr12_14720104_14720509 | 0.14 |
RP11-695J4.2 |
|
378 |
0.73 |
| chr9_138943188_138943742 | 0.14 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
1039 |
0.58 |
| chr5_149864820_149865733 | 0.14 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
105 |
0.97 |
| chr9_33523659_33524999 | 0.14 |
ANKRD18B |
ankyrin repeat domain 18B |
63 |
0.97 |
| chr4_119273101_119273655 | 0.14 |
PRSS12 |
protease, serine, 12 (neurotrypsin, motopsin) |
780 |
0.76 |
| chr15_96888963_96889465 | 0.14 |
ENSG00000222651 |
. |
12724 |
0.15 |
| chr9_25678314_25678778 | 0.14 |
TUSC1 |
tumor suppressor candidate 1 |
310 |
0.95 |
| chr9_112083182_112083638 | 0.13 |
EPB41L4B |
erythrocyte membrane protein band 4.1 like 4B |
166 |
0.95 |
| chr5_2756564_2757124 | 0.13 |
C5orf38 |
chromosome 5 open reading frame 38 |
4465 |
0.28 |
| chr2_79739509_79740065 | 0.13 |
CTNNA2 |
catenin (cadherin-associated protein), alpha 2 |
339 |
0.91 |
| chr20_9048816_9049419 | 0.13 |
PLCB4 |
phospholipase C, beta 4 |
293 |
0.9 |
| chr2_121499238_121499672 | 0.13 |
GLI2 |
GLI family zinc finger 2 |
5632 |
0.32 |
| chr9_133813639_133813991 | 0.13 |
FIBCD1 |
fibrinogen C domain containing 1 |
422 |
0.82 |
| chr17_72839202_72839484 | 0.13 |
GRIN2C |
glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
16655 |
0.11 |
| chr5_38846105_38846894 | 0.13 |
OSMR |
oncostatin M receptor |
398 |
0.91 |
| chr19_45350061_45350833 | 0.13 |
PVRL2 |
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
664 |
0.57 |
| chr18_77547944_77548277 | 0.13 |
KCNG2 |
potassium voltage-gated channel, subfamily G, member 2 |
75558 |
0.1 |
| chr1_159750747_159751482 | 0.13 |
DUSP23 |
dual specificity phosphatase 23 |
321 |
0.85 |
| chr3_39093427_39094272 | 0.13 |
WDR48 |
WD repeat domain 48 |
57 |
0.97 |
| chr12_124155530_124156430 | 0.13 |
TCTN2 |
tectonic family member 2 |
320 |
0.85 |
| chr1_119530163_119530409 | 0.13 |
TBX15 |
T-box 15 |
142 |
0.97 |
| chr11_86382517_86383664 | 0.13 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
150 |
0.98 |
| chr9_110250307_110251795 | 0.13 |
KLF4 |
Kruppel-like factor 4 (gut) |
360 |
0.89 |
| chr13_52979416_52980324 | 0.13 |
THSD1 |
thrombospondin, type I, domain containing 1 |
393 |
0.85 |
| chrX_19140154_19140583 | 0.13 |
GPR64 |
G protein-coupled receptor 64 |
211 |
0.97 |
| chr7_131240036_131240570 | 0.13 |
PODXL |
podocalyxin-like |
1063 |
0.65 |
| chr10_79396627_79397649 | 0.13 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
67 |
0.98 |
| chr5_131745958_131747299 | 0.13 |
C5orf56 |
chromosome 5 open reading frame 56 |
45 |
0.96 |
| chr17_1936770_1936951 | 0.13 |
DPH1 |
diphthamide biosynthesis 1 |
173 |
0.88 |
| chr11_128494350_128494501 | 0.13 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
36972 |
0.16 |
| chr5_176921947_176922464 | 0.13 |
RP11-1334A24.6 |
|
209 |
0.86 |
| chrX_37208613_37209248 | 0.13 |
PRRG1 |
proline rich Gla (G-carboxyglutamic acid) 1 |
298 |
0.54 |
| chr22_41843038_41844299 | 0.13 |
TOB2 |
transducer of ERBB2, 2 |
641 |
0.64 |
| chr8_56851767_56852412 | 0.13 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
704 |
0.63 |
| chr18_74843262_74844680 | 0.13 |
MBP |
myelin basic protein |
331 |
0.94 |
| chr7_45002030_45002849 | 0.13 |
RP4-647J21.1 |
|
1931 |
0.25 |
| chr1_1207989_1209115 | 0.13 |
UBE2J2 |
ubiquitin-conjugating enzyme E2, J2 |
299 |
0.76 |
| chr5_136834312_136835008 | 0.13 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
328 |
0.92 |
| chr6_5084934_5086227 | 0.13 |
PPP1R3G |
protein phosphatase 1, regulatory subunit 3G |
140 |
0.97 |
| chr3_147128128_147129466 | 0.13 |
ZIC1 |
Zic family member 1 |
1626 |
0.36 |
| chr1_30603166_30603378 | 0.13 |
ENSG00000222787 |
. |
245523 |
0.02 |
| chr11_61595087_61595308 | 0.13 |
FADS2 |
fatty acid desaturase 2 |
36 |
0.95 |
| chr13_96204772_96205414 | 0.13 |
CLDN10 |
claudin 10 |
114 |
0.97 |
| chr22_50781024_50781650 | 0.12 |
PPP6R2 |
protein phosphatase 6, regulatory subunit 2 |
396 |
0.74 |
| chr19_49617571_49617790 | 0.12 |
LIN7B |
lin-7 homolog B (C. elegans) |
1 |
0.94 |
| chr10_49514049_49514506 | 0.12 |
MAPK8 |
mitogen-activated protein kinase 8 |
421 |
0.88 |
| chr19_21769065_21769964 | 0.12 |
RP11-678G14.2 |
|
17475 |
0.22 |
| chr21_40984409_40984913 | 0.12 |
C21orf88 |
chromosome 21 open reading frame 88 |
87 |
0.97 |
| chr3_49055259_49056395 | 0.12 |
DALRD3 |
DALR anticodon binding domain containing 3 |
170 |
0.84 |
| chr18_2906194_2907483 | 0.12 |
RP11-737O24.5 |
|
14126 |
0.16 |
| chr2_177502027_177502943 | 0.12 |
ENSG00000252027 |
. |
26919 |
0.25 |
| chr3_120004025_120004467 | 0.12 |
GPR156 |
G protein-coupled receptor 156 |
305 |
0.92 |
| chr19_33790953_33791293 | 0.12 |
CTD-2540B15.11 |
|
283 |
0.79 |
| chr8_99439239_99439631 | 0.12 |
KCNS2 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
185 |
0.95 |
| chr5_132158871_132159069 | 0.12 |
SHROOM1 |
shroom family member 1 |
2862 |
0.16 |
| chr1_115397478_115398199 | 0.12 |
SYCP1 |
synaptonemal complex protein 1 |
84 |
0.98 |
| chr5_114193628_114194015 | 0.12 |
RP11-492A10.1 |
|
189714 |
0.03 |
| chr7_5467738_5468320 | 0.12 |
TNRC18 |
trinucleotide repeat containing 18 |
2984 |
0.19 |
| chr19_45147190_45148055 | 0.12 |
PVR |
poliovirus receptor |
225 |
0.9 |
| chr2_236578912_236579455 | 0.12 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
935 |
0.7 |
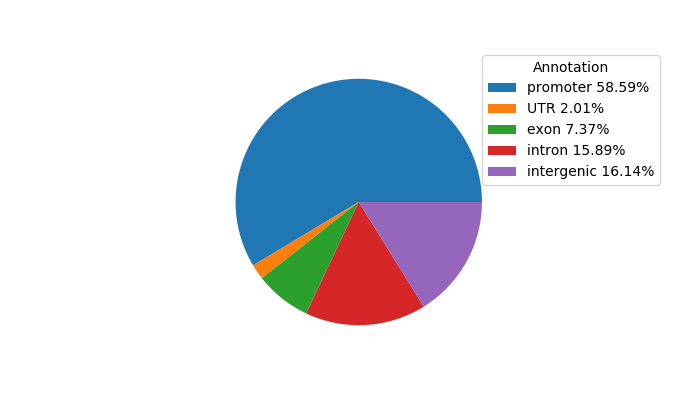
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.1 | 0.3 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.3 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.2 | GO:0060453 | regulation of gastric acid secretion(GO:0060453) |
| 0.1 | 0.1 | GO:0003079 | obsolete positive regulation of natriuresis(GO:0003079) |
| 0.1 | 0.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.1 | 0.1 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.2 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 0.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.2 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.1 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.2 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.2 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.3 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.2 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.2 | GO:0048343 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.7 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.0 | 0.1 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0060510 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0090026 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.0 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.0 | 0.0 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0010560 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0010658 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.2 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.0 | GO:0072193 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0043096 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.0 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.0 | GO:0042663 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.2 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.0 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.0 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.0 | GO:0003309 | type B pancreatic cell differentiation(GO:0003309) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0043302 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0006837 | serotonin transport(GO:0006837) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.0 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.0 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.3 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.0 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0031082 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.2 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0002060 | nucleobase binding(GO:0002054) purine nucleobase binding(GO:0002060) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.0 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0001077 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | Genes involved in Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |