Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
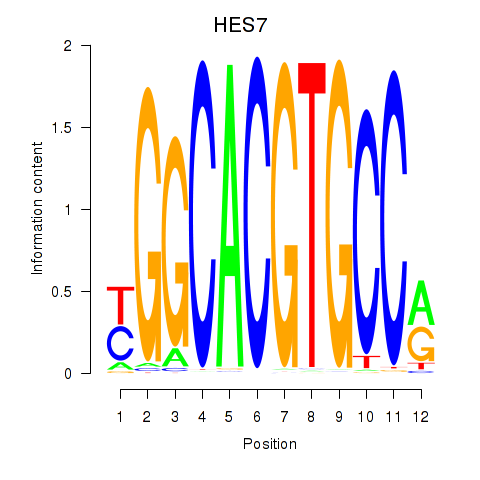
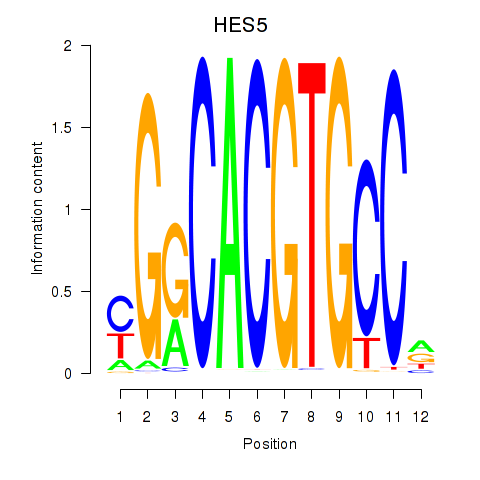
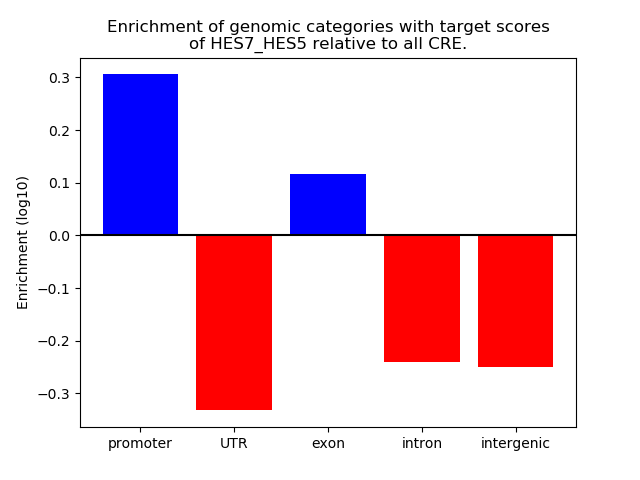
Results for HES7_HES5
Z-value: 0.69
Transcription factors associated with HES7_HES5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HES7
|
ENSG00000179111.4 | hes family bHLH transcription factor 7 |
|
HES5
|
ENSG00000197921.5 | hes family bHLH transcription factor 5 |
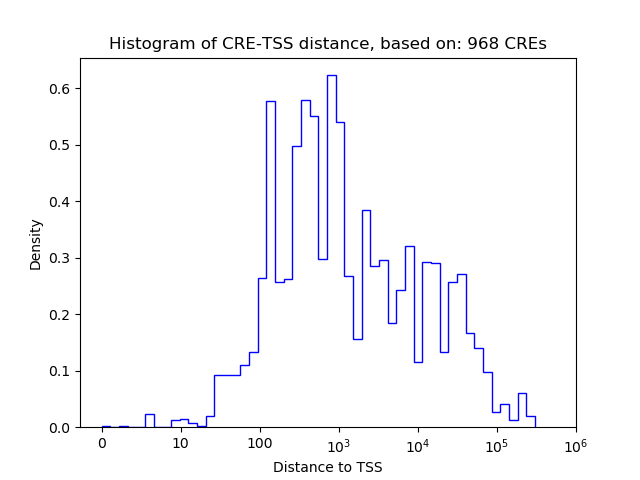
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_2467250_2467401 | HES5 | 5641 | 0.102622 | 0.41 | 2.7e-01 | Click! |
| chr1_2461361_2461622 | HES5 | 193 | 0.883039 | 0.33 | 3.9e-01 | Click! |
| chr1_2462303_2462454 | HES5 | 694 | 0.498546 | 0.31 | 4.1e-01 | Click! |
| chr1_2460280_2460431 | HES5 | 1329 | 0.250275 | 0.29 | 4.5e-01 | Click! |
| chr1_2460577_2460728 | HES5 | 1032 | 0.331461 | 0.12 | 7.5e-01 | Click! |
| chr17_8029184_8029766 | HES7 | 2065 | 0.148924 | 0.22 | 5.7e-01 | Click! |
| chr17_8025015_8025782 | HES7 | 2004 | 0.149938 | 0.21 | 5.9e-01 | Click! |
| chr17_8025785_8026039 | HES7 | 1490 | 0.202056 | -0.21 | 6.0e-01 | Click! |
| chr17_8028639_8028790 | HES7 | 1304 | 0.235636 | 0.19 | 6.3e-01 | Click! |
| chr17_8026048_8026346 | HES7 | 1205 | 0.255059 | -0.16 | 6.8e-01 | Click! |
Activity of the HES7_HES5 motif across conditions
Conditions sorted by the z-value of the HES7_HES5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
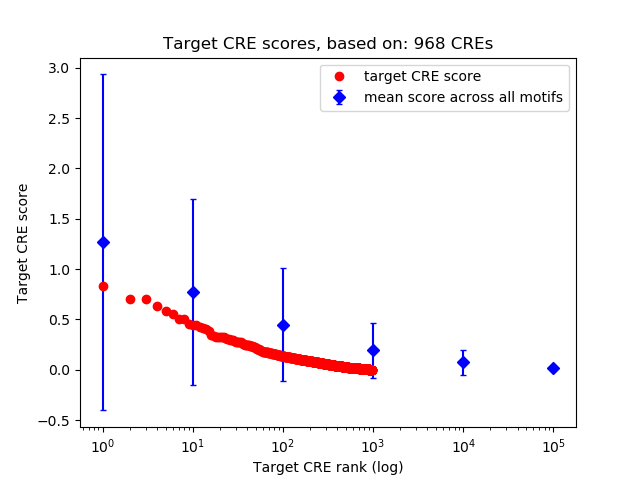
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_9714142_9714379 | 0.84 |
C1orf200 |
chromosome 1 open reading frame 200 |
384 |
0.82 |
| chr2_158295494_158296053 | 0.71 |
CYTIP |
cytohesin 1 interacting protein |
153 |
0.96 |
| chr11_2291181_2291518 | 0.70 |
ASCL2 |
achaete-scute family bHLH transcription factor 2 |
833 |
0.53 |
| chr14_105527312_105527781 | 0.63 |
GPR132 |
G protein-coupled receptor 132 |
3727 |
0.23 |
| chr1_154378708_154379562 | 0.58 |
RP11-350G8.5 |
|
95 |
0.93 |
| chr7_644134_644285 | 0.55 |
AC147651.4 |
|
1387 |
0.35 |
| chr3_196367453_196367902 | 0.51 |
PIGX |
phosphatidylinositol glycan anchor biosynthesis, class X |
1031 |
0.33 |
| chr21_34395985_34396161 | 0.50 |
OLIG2 |
oligodendrocyte lineage transcription factor 2 |
2080 |
0.33 |
| chr9_132647660_132648047 | 0.45 |
FNBP1 |
formin binding protein 1 |
33736 |
0.12 |
| chr19_18507519_18507670 | 0.44 |
LRRC25 |
leucine rich repeat containing 25 |
827 |
0.4 |
| chr20_55965895_55966111 | 0.44 |
RBM38 |
RNA binding motif protein 38 |
460 |
0.74 |
| chr7_157368911_157369331 | 0.43 |
ENSG00000207960 |
. |
2007 |
0.35 |
| chr2_68589306_68589626 | 0.41 |
PLEK |
pleckstrin |
2839 |
0.22 |
| chr1_160678982_160679166 | 0.41 |
CD48 |
CD48 molecule |
2519 |
0.24 |
| chr11_48029600_48029751 | 0.39 |
AC103828.1 |
|
7732 |
0.21 |
| chrX_56839719_56839908 | 0.35 |
ENSG00000204272 |
. |
84121 |
0.1 |
| chr2_202129112_202129390 | 0.33 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
2572 |
0.28 |
| chr21_45773621_45773812 | 0.33 |
TRPM2 |
transient receptor potential cation channel, subfamily M, member 2 |
145 |
0.91 |
| chr19_890673_890824 | 0.33 |
MED16 |
mediator complex subunit 16 |
383 |
0.68 |
| chr1_226250996_226251291 | 0.32 |
H3F3A |
H3 histone, family 3A |
535 |
0.7 |
| chr7_75622802_75623087 | 0.32 |
POR |
P450 (cytochrome) oxidoreductase |
11715 |
0.16 |
| chr13_30120185_30120336 | 0.32 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
40669 |
0.18 |
| chr4_18023348_18023557 | 0.32 |
LCORL |
ligand dependent nuclear receptor corepressor-like |
31 |
0.99 |
| chr19_19738537_19738987 | 0.31 |
LPAR2 |
lysophosphatidic acid receptor 2 |
153 |
0.92 |
| chr2_27487055_27487432 | 0.30 |
SLC30A3 |
solute carrier family 30 (zinc transporter), member 3 |
239 |
0.85 |
| chr15_75471070_75471460 | 0.30 |
C15orf39 |
chromosome 15 open reading frame 39 |
16719 |
0.13 |
| chr22_20785488_20785753 | 0.30 |
SCARF2 |
scavenger receptor class F, member 2 |
6492 |
0.12 |
| chr1_154976315_154976466 | 0.29 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
1095 |
0.25 |
| chr19_13928529_13928680 | 0.28 |
ZSWIM4 |
zinc finger, SWIM-type containing 4 |
91 |
0.92 |
| chr10_71905035_71905382 | 0.28 |
TYSND1 |
trypsin domain containing 1 |
1134 |
0.47 |
| chr11_65153709_65153977 | 0.28 |
FRMD8 |
FERM domain containing 8 |
227 |
0.87 |
| chr2_61112543_61112833 | 0.28 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
3897 |
0.23 |
| chr19_35838935_35839086 | 0.28 |
ENSG00000263397 |
. |
2594 |
0.13 |
| chr16_87888685_87888836 | 0.27 |
SLC7A5 |
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
8485 |
0.17 |
| chr12_102165709_102165860 | 0.26 |
ENSG00000222255 |
. |
6497 |
0.11 |
| chr16_89042817_89042968 | 0.26 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
509 |
0.75 |
| chr8_28259515_28259714 | 0.25 |
ZNF395 |
zinc finger protein 395 |
604 |
0.66 |
| chr16_48419475_48419990 | 0.25 |
SIAH1 |
siah E3 ubiquitin protein ligase 1 |
371 |
0.85 |
| chr12_663051_663343 | 0.25 |
B4GALNT3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
10898 |
0.16 |
| chr20_49011186_49011337 | 0.24 |
ENSG00000244376 |
. |
34759 |
0.19 |
| chr9_92099141_92099337 | 0.24 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
4434 |
0.26 |
| chr6_133134294_133134903 | 0.24 |
RPS12 |
ribosomal protein S12 |
982 |
0.36 |
| chr7_130597934_130598170 | 0.24 |
ENSG00000226380 |
. |
35754 |
0.19 |
| chr2_192306449_192306600 | 0.23 |
MYO1B |
myosin IB |
30733 |
0.22 |
| chr17_40440858_40441114 | 0.23 |
STAT5A |
signal transducer and activator of transcription 5A |
425 |
0.75 |
| chr14_104604081_104604495 | 0.23 |
CTD-2223O18.1 |
|
53 |
0.93 |
| chr12_4381475_4382675 | 0.23 |
CCND2 |
cyclin D2 |
863 |
0.52 |
| chr20_826059_826564 | 0.23 |
FAM110A |
family with sequence similarity 110, member A |
544 |
0.82 |
| chr15_81316371_81316635 | 0.22 |
C15orf26 |
chromosome 15 open reading frame 26 |
17129 |
0.15 |
| chr10_131908264_131908714 | 0.22 |
GLRX3 |
glutaredoxin 3 |
26174 |
0.26 |
| chr9_136727740_136727891 | 0.21 |
SARDH |
sarcosine dehydrogenase |
122738 |
0.05 |
| chr22_23527376_23527533 | 0.21 |
BCR |
breakpoint cluster region |
4902 |
0.16 |
| chr2_102974021_102974195 | 0.20 |
IL18R1 |
interleukin 18 receptor 1 |
1719 |
0.35 |
| chr19_17828394_17828681 | 0.20 |
MAP1S |
microtubule-associated protein 1S |
1514 |
0.3 |
| chr16_88987498_88987922 | 0.20 |
RP11-830F9.7 |
|
16454 |
0.11 |
| chr6_29911146_29911299 | 0.20 |
HLA-A |
major histocompatibility complex, class I, A |
896 |
0.59 |
| chr1_192777226_192777377 | 0.19 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
870 |
0.71 |
| chr13_34116896_34117047 | 0.19 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
192204 |
0.03 |
| chr9_36142717_36143534 | 0.18 |
GLIPR2 |
GLI pathogenesis-related 2 |
6383 |
0.2 |
| chr19_6425590_6425914 | 0.18 |
KHSRP |
KH-type splicing regulatory protein |
947 |
0.34 |
| chr12_108056413_108056564 | 0.18 |
ENSG00000222160 |
. |
11922 |
0.16 |
| chr9_136602785_136602936 | 0.18 |
SARDH |
sarcosine dehydrogenase |
617 |
0.78 |
| chr10_28622520_28622887 | 0.18 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
712 |
0.72 |
| chr10_103539328_103539744 | 0.18 |
NPM3 |
nucleophosmin/nucleoplasmin 3 |
3634 |
0.18 |
| chr7_26191850_26192061 | 0.17 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
95 |
0.97 |
| chr3_176919226_176919959 | 0.17 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
4331 |
0.31 |
| chr1_226645087_226645238 | 0.17 |
ENSG00000223282 |
. |
11524 |
0.22 |
| chr6_117869124_117869373 | 0.17 |
GOPC |
golgi-associated PDZ and coiled-coil motif containing |
54279 |
0.12 |
| chr17_40439619_40439799 | 0.17 |
STAT5A |
signal transducer and activator of transcription 5A |
144 |
0.93 |
| chrX_70401472_70401874 | 0.17 |
RP5-1091N2.9 |
|
16352 |
0.12 |
| chr19_58898175_58898426 | 0.17 |
ENSG00000266640 |
. |
75 |
0.77 |
| chr4_140476590_140476990 | 0.17 |
SETD7 |
SET domain containing (lysine methyltransferase) 7 |
572 |
0.65 |
| chr15_41804782_41804942 | 0.17 |
LTK |
leukocyte receptor tyrosine kinase |
1132 |
0.4 |
| chr16_1980035_1980237 | 0.16 |
HS3ST6 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
11695 |
0.06 |
| chr2_43452270_43452631 | 0.16 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
1298 |
0.5 |
| chr1_160316893_160317044 | 0.16 |
NCSTN |
nicastrin |
2484 |
0.17 |
| chr2_190442035_190442186 | 0.16 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
3503 |
0.3 |
| chr1_27668600_27669070 | 0.16 |
SYTL1 |
synaptotagmin-like 1 |
322 |
0.82 |
| chr10_6018795_6019199 | 0.16 |
IL15RA |
interleukin 15 receptor, alpha |
180 |
0.93 |
| chr6_99282314_99282478 | 0.16 |
POU3F2 |
POU class 3 homeobox 2 |
184 |
0.97 |
| chr3_133292516_133292718 | 0.15 |
CDV3 |
CDV3 homolog (mouse) |
43 |
0.98 |
| chr14_51296169_51297325 | 0.15 |
NIN |
ninein (GSK3B interacting protein) |
450 |
0.76 |
| chr16_81129212_81129933 | 0.15 |
GCSH |
glycine cleavage system protein H (aminomethyl carrier) |
273 |
0.86 |
| chr2_25149877_25150037 | 0.15 |
ADCY3 |
adenylate cyclase 3 |
7249 |
0.18 |
| chr6_111197567_111198255 | 0.15 |
AMD1 |
adenosylmethionine decarboxylase 1 |
1133 |
0.45 |
| chr7_74988426_74988792 | 0.15 |
STAG3L1 |
stromal antigen 3-like 1 (pseudogene) |
154 |
0.91 |
| chr4_154217109_154217260 | 0.15 |
ENSG00000201786 |
. |
29022 |
0.14 |
| chr1_25944160_25944528 | 0.15 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
3 |
0.98 |
| chr18_43431286_43431437 | 0.15 |
SIGLEC15 |
sialic acid binding Ig-like lectin 15 |
13555 |
0.2 |
| chr16_85089458_85089881 | 0.15 |
KIAA0513 |
KIAA0513 |
7149 |
0.23 |
| chr4_113153042_113153239 | 0.15 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
151 |
0.76 |
| chr7_66767653_66767865 | 0.15 |
PMS2P4 |
postmeiotic segregation increased 2 pseudogene 4 |
135 |
0.95 |
| chr11_64108635_64108896 | 0.14 |
CCDC88B |
coiled-coil domain containing 88B |
1070 |
0.28 |
| chr3_156391824_156392138 | 0.14 |
TIPARP |
TCDD-inducible poly(ADP-ribose) polymerase |
150 |
0.55 |
| chr22_37881004_37881696 | 0.14 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
683 |
0.62 |
| chr21_43933401_43933705 | 0.14 |
SLC37A1 |
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
471 |
0.74 |
| chr11_93394564_93394780 | 0.14 |
KIAA1731 |
KIAA1731 |
133 |
0.94 |
| chr1_156426947_156427327 | 0.14 |
MEF2D |
myocyte enhancer factor 2D |
26085 |
0.1 |
| chr11_65185943_65186437 | 0.14 |
ENSG00000245532 |
. |
25739 |
0.09 |
| chr1_144521538_144522007 | 0.14 |
RP11-640M9.1 |
|
198 |
0.94 |
| chr7_5597597_5597858 | 0.13 |
CTB-161C1.1 |
|
1365 |
0.33 |
| chr9_92219727_92219878 | 0.13 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
126 |
0.98 |
| chr15_85523451_85523602 | 0.13 |
PDE8A |
phosphodiesterase 8A |
145 |
0.96 |
| chr16_85605071_85605483 | 0.13 |
GSE1 |
Gse1 coiled-coil protein |
39738 |
0.16 |
| chr15_101548752_101549003 | 0.13 |
RP11-505E24.3 |
|
40784 |
0.16 |
| chr17_42988666_42988862 | 0.13 |
GFAP |
glial fibrillary acidic protein |
745 |
0.46 |
| chr2_20849968_20850393 | 0.13 |
HS1BP3 |
HCLS1 binding protein 3 |
658 |
0.74 |
| chr14_23025391_23025542 | 0.13 |
AE000662.92 |
Uncharacterized protein |
68 |
0.87 |
| chr3_49026937_49027533 | 0.13 |
P4HTM |
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
84 |
0.63 |
| chrX_118827540_118827798 | 0.13 |
SEPT6 |
septin 6 |
336 |
0.88 |
| chr19_48103713_48103864 | 0.13 |
GLTSCR1 |
glioma tumor suppressor candidate region gene 1 |
7665 |
0.14 |
| chr2_198071025_198071176 | 0.13 |
ANKRD44 |
ankyrin repeat domain 44 |
8338 |
0.21 |
| chr8_145192644_145192918 | 0.13 |
FAM203A |
family with sequence similarity 203, member A |
109 |
0.93 |
| chr22_45634545_45634696 | 0.13 |
KIAA0930 |
KIAA0930 |
2030 |
0.3 |
| chr10_75531995_75532402 | 0.13 |
FUT11 |
fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
149 |
0.52 |
| chr1_143744977_143745422 | 0.13 |
PPIAL4G |
peptidylprolyl isomerase A (cyclophilin A)-like 4G |
22682 |
0.16 |
| chr6_13488651_13488802 | 0.13 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
832 |
0.5 |
| chr1_202611622_202611773 | 0.13 |
SYT2 |
synaptotagmin II |
884 |
0.64 |
| chr11_72853542_72853693 | 0.12 |
FCHSD2 |
FCH and double SH3 domains 2 |
311 |
0.91 |
| chr13_21050204_21050392 | 0.12 |
ENSG00000263978 |
. |
42313 |
0.15 |
| chr9_94186787_94187440 | 0.12 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
969 |
0.69 |
| chr19_917582_917891 | 0.12 |
KISS1R |
KISS1 receptor |
233 |
0.83 |
| chr10_129846734_129847253 | 0.12 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
1159 |
0.62 |
| chr6_143771128_143771800 | 0.12 |
ADAT2 |
adenosine deaminase, tRNA-specific 2 |
346 |
0.45 |
| chr8_28352405_28352827 | 0.12 |
FZD3 |
frizzled family receptor 3 |
843 |
0.51 |
| chr1_144341547_144341785 | 0.12 |
AL592284.1 |
Protein LOC642441 |
1928 |
0.34 |
| chr3_25825194_25825429 | 0.12 |
NGLY1 |
N-glycanase 1 |
269 |
0.8 |
| chr2_74648754_74648905 | 0.12 |
WDR54 |
WD repeat domain 54 |
27 |
0.58 |
| chr11_69489461_69490079 | 0.12 |
ORAOV1 |
oral cancer overexpressed 1 |
319 |
0.9 |
| chr16_68319324_68319536 | 0.12 |
ENSG00000252026 |
. |
1209 |
0.25 |
| chr1_147932755_147932982 | 0.12 |
PPIAL4A |
peptidylprolyl isomerase A (cyclophilin A)-like 4A |
22551 |
0.16 |
| chr3_135969527_135970046 | 0.12 |
PCCB |
propionyl CoA carboxylase, beta polypeptide |
299 |
0.94 |
| chr7_72476067_72476443 | 0.11 |
STAG3L3 |
stromal antigen 3-like 3 |
185 |
0.89 |
| chrX_53710808_53711038 | 0.11 |
HUWE1 |
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
191 |
0.97 |
| chr8_145114785_145114936 | 0.11 |
OPLAH |
5-oxoprolinase (ATP-hydrolysing) |
724 |
0.42 |
| chr5_49961781_49962268 | 0.11 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
184 |
0.97 |
| chr3_9885443_9885670 | 0.11 |
RPUSD3 |
RNA pseudouridylate synthase domain containing 3 |
70 |
0.94 |
| chr12_104964115_104964290 | 0.11 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
18424 |
0.22 |
| chr6_144536832_144537278 | 0.11 |
STX11 |
syntaxin 11 |
65392 |
0.12 |
| chr11_47416116_47416267 | 0.11 |
RP11-750H9.5 |
|
1000 |
0.33 |
| chr14_99726709_99727240 | 0.11 |
AL109767.1 |
|
2311 |
0.33 |
| chr1_145455050_145455272 | 0.11 |
POLR3GL |
polymerase (RNA) III (DNA directed) polypeptide G (32kD)-like |
15226 |
0.1 |
| chr7_101460813_101461304 | 0.11 |
CUX1 |
cut-like homeobox 1 |
138 |
0.97 |
| chr6_139456178_139456587 | 0.11 |
HECA |
headcase homolog (Drosophila) |
133 |
0.97 |
| chr8_74206830_74207171 | 0.11 |
RDH10 |
retinol dehydrogenase 10 (all-trans) |
153 |
0.72 |
| chr10_89823812_89823963 | 0.11 |
ENSG00000200891 |
. |
69435 |
0.12 |
| chr11_1874347_1875032 | 0.11 |
LSP1 |
lymphocyte-specific protein 1 |
489 |
0.65 |
| chr12_57080547_57081320 | 0.11 |
PTGES3 |
prostaglandin E synthase 3 (cytosolic) |
1013 |
0.41 |
| chr11_107992319_107993285 | 0.11 |
ACAT1 |
acetyl-CoA acetyltransferase 1 |
279 |
0.6 |
| chr22_40742518_40742799 | 0.11 |
ADSL |
adenylosuccinate lyase |
122 |
0.96 |
| chr17_80188177_80188492 | 0.11 |
SLC16A3 |
solute carrier family 16 (monocarboxylate transporter), member 3 |
1365 |
0.28 |
| chr3_38496543_38496888 | 0.11 |
ACVR2B-AS1 |
ACVR2B antisense RNA 1 |
404 |
0.78 |
| chr20_5588883_5589774 | 0.10 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
2344 |
0.4 |
| chr5_138724836_138724987 | 0.10 |
MZB1 |
marginal zone B and B1 cell-specific protein |
675 |
0.5 |
| chr19_14544413_14545000 | 0.10 |
PKN1 |
protein kinase N1 |
401 |
0.76 |
| chr9_71148060_71148211 | 0.10 |
TMEM252 |
transmembrane protein 252 |
7648 |
0.27 |
| chr19_8289482_8289633 | 0.10 |
CERS4 |
ceramide synthase 4 |
354 |
0.85 |
| chr22_36425098_36425275 | 0.10 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
713 |
0.78 |
| chr17_25958886_25959081 | 0.10 |
LGALS9 |
lectin, galactoside-binding, soluble, 9 |
750 |
0.6 |
| chr11_2324171_2324374 | 0.10 |
TSPAN32 |
tetraspanin 32 |
174 |
0.88 |
| chr11_121526465_121526781 | 0.10 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
65495 |
0.14 |
| chr18_74280302_74280453 | 0.10 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
39503 |
0.16 |
| chr1_46154130_46154324 | 0.10 |
TMEM69 |
transmembrane protein 69 |
1341 |
0.32 |
| chr1_148176495_148177244 | 0.10 |
PPIAL4D |
peptidylprolyl isomerase A (cyclophilin A)-like 4D |
25667 |
0.21 |
| chr10_99093753_99093904 | 0.10 |
RP11-452K12.4 |
|
493 |
0.49 |
| chr11_66405551_66405805 | 0.10 |
RBM4 |
RNA binding motif protein 4 |
410 |
0.71 |
| chr14_52784712_52784898 | 0.10 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
3692 |
0.31 |
| chr4_139936230_139936672 | 0.10 |
CCRN4L |
CCR4 carbon catabolite repression 4-like (S. cerevisiae) |
492 |
0.71 |
| chr7_150415780_150415950 | 0.10 |
GIMAP1 |
GTPase, IMAP family member 1 |
2220 |
0.26 |
| chr11_76869474_76869625 | 0.10 |
MYO7A |
myosin VIIA |
21322 |
0.2 |
| chr5_159894836_159895683 | 0.10 |
ENSG00000265237 |
. |
6150 |
0.17 |
| chr6_41908853_41909458 | 0.10 |
CCND3 |
cyclin D3 |
36 |
0.97 |
| chr16_67189130_67189364 | 0.10 |
TRADD |
TNFRSF1A-associated via death domain |
905 |
0.31 |
| chr8_145437857_145438162 | 0.10 |
FAM203B |
family with sequence similarity 203, member B |
129 |
0.94 |
| chr1_236445510_236445882 | 0.10 |
ERO1LB |
ERO1-like beta (S. cerevisiae) |
377 |
0.86 |
| chr2_152495160_152495321 | 0.09 |
NEB |
nebulin |
55107 |
0.16 |
| chr3_10183779_10183952 | 0.09 |
VHL |
von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase |
393 |
0.67 |
| chr13_41239028_41239270 | 0.09 |
FOXO1 |
forkhead box O1 |
1585 |
0.46 |
| chr7_5595191_5595362 | 0.09 |
CTB-161C1.1 |
|
1086 |
0.41 |
| chr17_40441446_40441597 | 0.09 |
STAT5A |
signal transducer and activator of transcription 5A |
960 |
0.43 |
| chr8_38240572_38240933 | 0.09 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
962 |
0.42 |
| chr1_894870_895078 | 0.09 |
NOC2L |
nucleolar complex associated 2 homolog (S. cerevisiae) |
304 |
0.72 |
| chr22_24038076_24038227 | 0.09 |
RGL4 |
ral guanine nucleotide dissociation stimulator-like 4 |
520 |
0.69 |
| chr5_118734822_118734973 | 0.09 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
43167 |
0.13 |
| chr3_37934486_37934637 | 0.09 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
30896 |
0.15 |
| chr1_173836617_173837079 | 0.09 |
ENSG00000234741 |
. |
40 |
0.83 |
| chr4_166128900_166129705 | 0.09 |
KLHL2 |
kelch-like family member 2 |
327 |
0.55 |
| chr7_99933726_99933950 | 0.09 |
PILRB |
paired immunoglobin-like type 2 receptor beta |
101 |
0.92 |
| chr18_33078024_33078260 | 0.09 |
INO80C |
INO80 complex subunit C |
187 |
0.57 |
| chr8_65711600_65711819 | 0.09 |
CYP7B1 |
cytochrome P450, family 7, subfamily B, polypeptide 1 |
391 |
0.91 |
| chr1_27019608_27020111 | 0.09 |
RP5-968P14.2 |
|
763 |
0.56 |
| chr12_25204572_25204723 | 0.09 |
LRMP |
lymphoid-restricted membrane protein |
558 |
0.78 |
| chr2_149401643_149402183 | 0.09 |
EPC2 |
enhancer of polycomb homolog 2 (Drosophila) |
96 |
0.98 |
| chr3_195634754_195635357 | 0.09 |
TNK2-AS1 |
TNK2 antisense RNA 1 |
108 |
0.91 |
| chr12_122249441_122249966 | 0.09 |
SETD1B |
SET domain containing 1B |
7065 |
0.16 |
| chr12_109024686_109024975 | 0.09 |
SELPLG |
selectin P ligand |
1024 |
0.4 |
| chr14_99741095_99741246 | 0.09 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
3309 |
0.27 |
| chr11_58345573_58345832 | 0.09 |
LPXN |
leupaxin |
9 |
0.95 |
| chr20_23066190_23066341 | 0.09 |
CD93 |
CD93 molecule |
712 |
0.66 |
| chr2_38871082_38871242 | 0.09 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
21890 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046449 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.1 | 0.2 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.1 | 0.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.1 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.0 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.2 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.2 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.3 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |