Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
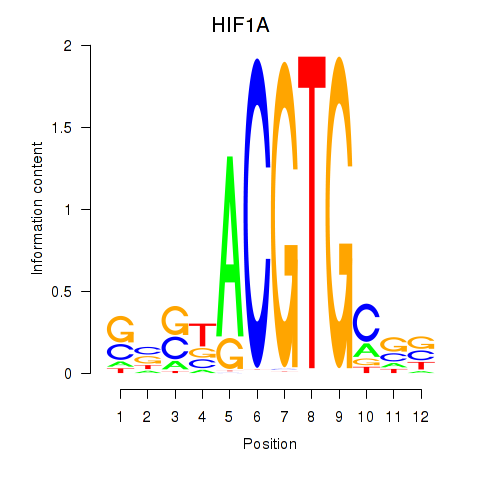
Results for HIF1A
Z-value: 0.62
Transcription factors associated with HIF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIF1A
|
ENSG00000100644.12 | hypoxia inducible factor 1 subunit alpha |
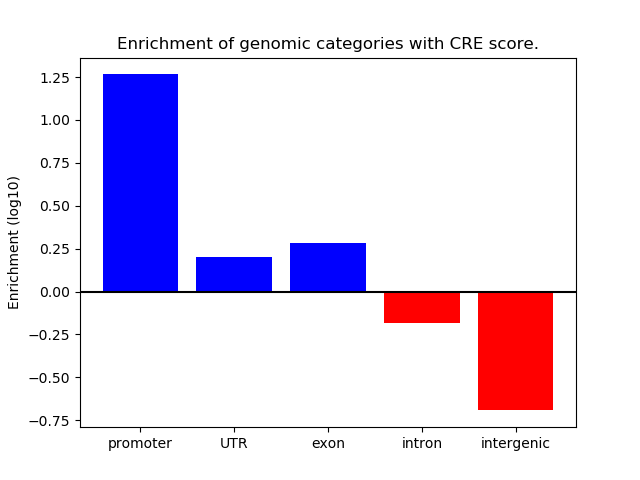
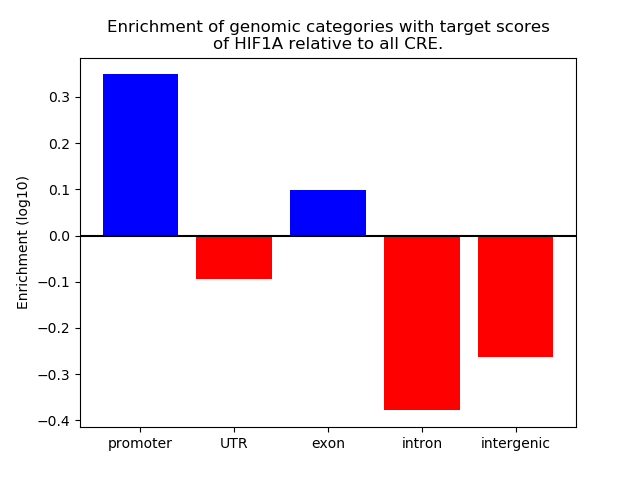
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_62168732_62168883 | HIF1A | 4467 | 0.276581 | 0.58 | 1.1e-01 | Click! |
| chr14_62171830_62171981 | HIF1A | 7565 | 0.249667 | 0.57 | 1.1e-01 | Click! |
| chr14_62185926_62186077 | HIF1A | 21661 | 0.210863 | 0.52 | 1.5e-01 | Click! |
| chr14_62144501_62144652 | HIF1A | 17682 | 0.211114 | -0.49 | 1.8e-01 | Click! |
| chr14_62144211_62144362 | HIF1A | 17972 | 0.210257 | -0.46 | 2.2e-01 | Click! |
Activity of the HIF1A motif across conditions
Conditions sorted by the z-value of the HIF1A motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
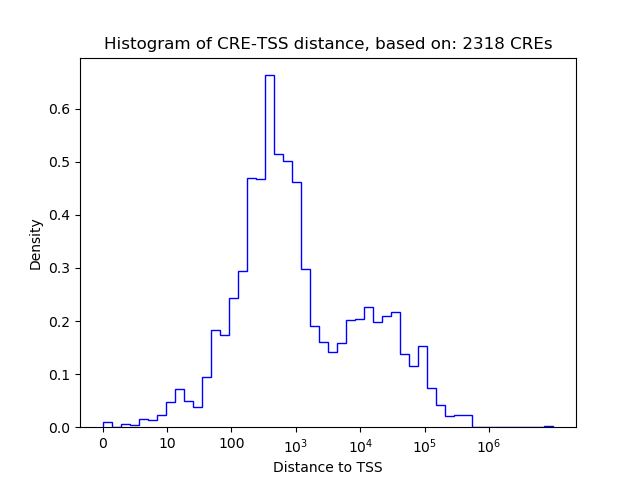
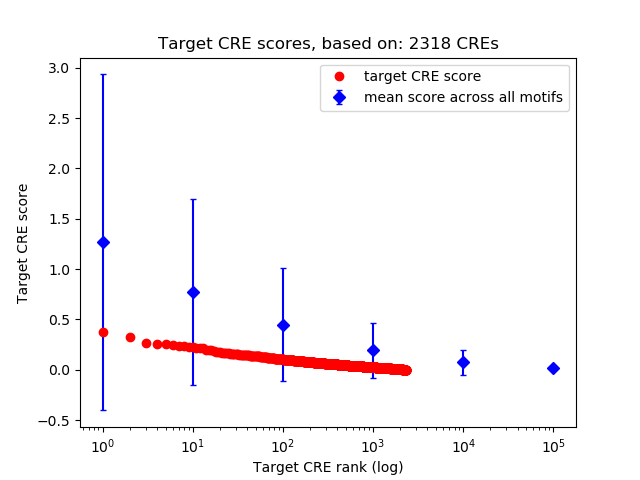
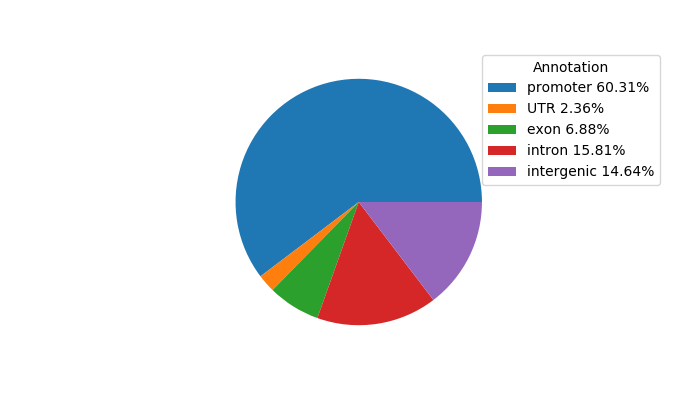
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_55087325_55087551 | 0.38 |
EGFR |
epidermal growth factor receptor |
627 |
0.83 |
| chr14_30396439_30396658 | 0.32 |
PRKD1 |
protein kinase D1 |
300 |
0.93 |
| chr4_17783154_17783359 | 0.27 |
FAM184B |
family with sequence similarity 184, member B |
121 |
0.97 |
| chr11_57406082_57406969 | 0.26 |
AP000662.4 |
|
676 |
0.5 |
| chr5_131991447_131991948 | 0.25 |
AC004041.2 |
|
113 |
0.94 |
| chr3_171918528_171919229 | 0.24 |
ENSG00000243398 |
. |
36238 |
0.18 |
| chr19_46916092_46916684 | 0.24 |
CCDC8 |
coiled-coil domain containing 8 |
453 |
0.75 |
| chr12_58120609_58120962 | 0.23 |
AGAP2-AS1 |
AGAP2 antisense RNA 1 |
731 |
0.39 |
| chr4_183064801_183065567 | 0.23 |
AC108142.1 |
|
143 |
0.83 |
| chr15_95869891_95870263 | 0.23 |
ENSG00000222076 |
. |
418956 |
0.01 |
| chr14_55119300_55119828 | 0.21 |
SAMD4A |
sterile alpha motif domain containing 4A |
84927 |
0.09 |
| chr5_169172915_169173066 | 0.21 |
DOCK2 |
dedicator of cytokinesis 2 |
13731 |
0.24 |
| chr5_83017716_83018280 | 0.21 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
566 |
0.78 |
| chr11_35640577_35641198 | 0.20 |
FJX1 |
four jointed box 1 (Drosophila) |
1152 |
0.57 |
| chr15_83875054_83875562 | 0.19 |
HDGFRP3 |
Hepatoma-derived growth factor-related protein 3 |
1462 |
0.4 |
| chr18_24764849_24765000 | 0.19 |
CHST9 |
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
338 |
0.94 |
| chr5_9546456_9546803 | 0.19 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
442 |
0.8 |
| chrX_107179250_107179655 | 0.18 |
RP6-191P20.4 |
|
242 |
0.93 |
| chr17_59476045_59476898 | 0.17 |
RP11-332H18.4 |
|
260 |
0.74 |
| chr5_172754207_172754714 | 0.17 |
STC2 |
stanniocalcin 2 |
596 |
0.75 |
| chr9_101983648_101983918 | 0.17 |
ALG2 |
ALG2, alpha-1,3/1,6-mannosyltransferase |
454 |
0.53 |
| chr3_141088214_141088426 | 0.17 |
RP11-438D8.2 |
|
2341 |
0.33 |
| chr15_23931247_23932124 | 0.17 |
NDN |
necdin, melanoma antigen (MAGE) family member |
765 |
0.7 |
| chr10_122216113_122216362 | 0.17 |
PPAPDC1A |
phosphatidic acid phosphatase type 2 domain containing 1A |
229 |
0.95 |
| chr17_7342924_7343492 | 0.16 |
FGF11 |
fibroblast growth factor 11 |
9 |
0.91 |
| chr4_93226727_93227337 | 0.16 |
GRID2 |
glutamate receptor, ionotropic, delta 2 |
1224 |
0.46 |
| chrX_54385157_54385424 | 0.16 |
WNK3 |
WNK lysine deficient protein kinase 3 |
215 |
0.95 |
| chr19_13135793_13135990 | 0.16 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
80 |
0.94 |
| chrX_105969921_105970262 | 0.15 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
197 |
0.96 |
| chr14_64804508_64805275 | 0.15 |
ESR2 |
estrogen receptor 2 (ER beta) |
61 |
0.87 |
| chr20_4129930_4130735 | 0.15 |
SMOX |
spermine oxidase |
750 |
0.71 |
| chr3_159482295_159482916 | 0.15 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
287 |
0.9 |
| chr16_56660383_56660534 | 0.15 |
MT1E |
metallothionein 1E |
748 |
0.38 |
| chr6_138571879_138572276 | 0.15 |
PBOV1 |
prostate and breast cancer overexpressed 1 |
32450 |
0.2 |
| chr10_13931045_13931263 | 0.15 |
FRMD4A |
FERM domain containing 4A |
30242 |
0.2 |
| chr14_69013839_69014176 | 0.15 |
CTD-2325P2.4 |
|
81155 |
0.1 |
| chrX_48930077_48930476 | 0.15 |
PRAF2 |
PRA1 domain family, member 2 |
1372 |
0.22 |
| chr13_37494481_37494632 | 0.15 |
SMAD9 |
SMAD family member 9 |
147 |
0.96 |
| chr10_16562894_16563258 | 0.15 |
C1QL3 |
complement component 1, q subcomponent-like 3 |
928 |
0.63 |
| chr4_23890359_23890765 | 0.14 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
1096 |
0.66 |
| chr4_115519485_115519837 | 0.14 |
UGT8 |
UDP glycosyltransferase 8 |
50 |
0.99 |
| chr7_15725092_15725754 | 0.14 |
MEOX2 |
mesenchyme homeobox 2 |
1014 |
0.63 |
| chr3_133646028_133646432 | 0.14 |
C3orf36 |
chromosome 3 open reading frame 36 |
2426 |
0.34 |
| chr20_11138132_11138283 | 0.14 |
C20orf187 |
chromosome 20 open reading frame 187 |
129396 |
0.06 |
| chr9_133884490_133884806 | 0.14 |
LAMC3 |
laminin, gamma 3 |
179 |
0.95 |
| chr3_170137410_170137695 | 0.14 |
CLDN11 |
claudin 11 |
697 |
0.77 |
| chr11_2187699_2187850 | 0.14 |
TH |
tyrosine hydroxylase |
1562 |
0.23 |
| chr7_43152327_43152563 | 0.14 |
HECW1 |
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
201 |
0.93 |
| chr10_49731531_49732112 | 0.14 |
ARHGAP22 |
Rho GTPase activating protein 22 |
460 |
0.85 |
| chr11_119455314_119455975 | 0.14 |
RP11-196E1.3 |
|
23491 |
0.19 |
| chr1_158968705_158968932 | 0.14 |
IFI16 |
interferon, gamma-inducible protein 16 |
940 |
0.6 |
| chr19_49575534_49575685 | 0.14 |
KCNA7 |
potassium voltage-gated channel, shaker-related subfamily, member 7 |
589 |
0.44 |
| chr6_7909475_7910402 | 0.13 |
TXNDC5 |
thioredoxin domain containing 5 (endoplasmic reticulum) |
372 |
0.92 |
| chr6_169653425_169653684 | 0.13 |
THBS2 |
thrombospondin 2 |
585 |
0.83 |
| chr1_16399640_16399936 | 0.13 |
FAM131C |
family with sequence similarity 131, member C |
333 |
0.84 |
| chr15_91500342_91500803 | 0.13 |
RCCD1 |
RCC1 domain containing 1 |
1546 |
0.18 |
| chr18_8706613_8707165 | 0.13 |
SOGA2 |
SOGA family member 2 |
520 |
0.55 |
| chr7_94536809_94537187 | 0.13 |
PPP1R9A |
protein phosphatase 1, regulatory subunit 9A |
11 |
0.99 |
| chr19_55591836_55592067 | 0.13 |
EPS8L1 |
EPS8-like 1 |
191 |
0.89 |
| chr5_81701799_81702115 | 0.13 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
100791 |
0.08 |
| chr19_53561526_53561765 | 0.13 |
ERVV-2 |
endogenous retrovirus group V, member 2 |
13654 |
0.13 |
| chr8_37823203_37823407 | 0.13 |
ADRB3 |
adrenoceptor beta 3 |
1178 |
0.39 |
| chr3_62359874_62360076 | 0.13 |
FEZF2 |
FEZ family zinc finger 2 |
24 |
0.98 |
| chr14_75389218_75389970 | 0.13 |
RPS6KL1 |
ribosomal protein S6 kinase-like 1 |
360 |
0.84 |
| chr9_22005843_22006115 | 0.12 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
2973 |
0.2 |
| chr16_22826079_22826361 | 0.12 |
HS3ST2 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
722 |
0.79 |
| chr7_28076392_28076791 | 0.12 |
JAZF1 |
JAZF zinc finger 1 |
16173 |
0.26 |
| chr22_43115644_43116713 | 0.12 |
A4GALT |
alpha 1,4-galactosyltransferase |
24539 |
0.15 |
| chr20_19738276_19738696 | 0.12 |
AL121761.2 |
Uncharacterized protein |
193 |
0.95 |
| chr16_21312757_21313165 | 0.12 |
CRYM |
crystallin, mu |
1411 |
0.46 |
| chr22_44433690_44434013 | 0.12 |
PARVB |
parvin, beta |
6606 |
0.26 |
| chr7_47576453_47576928 | 0.12 |
TNS3 |
tensin 3 |
2185 |
0.45 |
| chr1_1490221_1490372 | 0.12 |
TMEM240 |
transmembrane protein 240 |
14463 |
0.09 |
| chr9_8733649_8733843 | 0.12 |
PTPRD |
protein tyrosine phosphatase, receptor type, D |
148 |
0.97 |
| chr15_73976688_73977423 | 0.12 |
CD276 |
CD276 molecule |
17 |
0.98 |
| chr2_74968036_74968578 | 0.11 |
SEMA4F |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
86869 |
0.07 |
| chr11_60609533_60610326 | 0.11 |
CCDC86 |
coiled-coil domain containing 86 |
385 |
0.51 |
| chr8_42356526_42357223 | 0.11 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
1887 |
0.34 |
| chr5_38846105_38846894 | 0.11 |
OSMR |
oncostatin M receptor |
398 |
0.91 |
| chr1_16275488_16275712 | 0.11 |
ZBTB17 |
zinc finger and BTB domain containing 17 |
4677 |
0.16 |
| chr17_1665908_1666244 | 0.11 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
47 |
0.96 |
| chr7_116660327_116660575 | 0.11 |
ST7 |
suppression of tumorigenicity 7 |
120 |
0.96 |
| chr20_53092350_53092560 | 0.11 |
DOK5 |
docking protein 5 |
198 |
0.97 |
| chr11_133907029_133907299 | 0.11 |
JAM3 |
junctional adhesion molecule 3 |
31656 |
0.18 |
| chr7_56183478_56183932 | 0.11 |
NUPR1L |
nuclear protein, transcriptional regulator, 1-like |
388 |
0.78 |
| chr11_19367315_19367499 | 0.11 |
NAV2 |
neuron navigator 2 |
4864 |
0.23 |
| chr6_1381911_1382148 | 0.11 |
RP4-668J24.2 |
|
3272 |
0.28 |
| chr15_83316430_83317127 | 0.11 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
50 |
0.76 |
| chr8_15095014_15095265 | 0.11 |
SGCZ |
sarcoglycan, zeta |
709 |
0.82 |
| chr2_239072349_239072829 | 0.11 |
FAM132B |
family with sequence similarity 132, member B |
385 |
0.8 |
| chr11_33397614_33397834 | 0.11 |
ENSG00000223134 |
. |
21713 |
0.24 |
| chr20_13976647_13977145 | 0.11 |
SEL1L2 |
sel-1 suppressor of lin-12-like 2 (C. elegans) |
193 |
0.78 |
| chr19_1241899_1242560 | 0.11 |
ATP5D |
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
410 |
0.68 |
| chr4_142053459_142053770 | 0.10 |
RNF150 |
ring finger protein 150 |
382 |
0.92 |
| chr6_54564223_54564374 | 0.10 |
ENSG00000251946 |
. |
86887 |
0.1 |
| chr5_122425011_122425806 | 0.10 |
AC106786.1 |
|
418 |
0.57 |
| chr1_17019431_17019582 | 0.10 |
ENSG00000264742 |
. |
11756 |
0.12 |
| chr18_43913317_43913996 | 0.10 |
RNF165 |
ring finger protein 165 |
275 |
0.95 |
| chr20_33300382_33300533 | 0.10 |
TP53INP2 |
tumor protein p53 inducible nuclear protein 2 |
7918 |
0.22 |
| chr10_5708404_5708555 | 0.10 |
ASB13 |
ankyrin repeat and SOCS box containing 13 |
62 |
0.97 |
| chr7_41736310_41736535 | 0.10 |
INHBA-AS1 |
INHBA antisense RNA 1 |
2876 |
0.26 |
| chr1_214156478_214156705 | 0.10 |
PROX1 |
prospero homeobox 1 |
67 |
0.98 |
| chr19_290704_290961 | 0.10 |
PPAP2C |
phosphatidic acid phosphatase type 2C |
338 |
0.87 |
| chr4_8271771_8271998 | 0.10 |
HTRA3 |
HtrA serine peptidase 3 |
380 |
0.89 |
| chr20_19956104_19956326 | 0.10 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
41545 |
0.15 |
| chr19_44951986_44952721 | 0.10 |
ZNF229 |
zinc finger protein 229 |
296 |
0.89 |
| chr13_20751391_20751542 | 0.10 |
LINC00556 |
long intergenic non-protein coding RNA 556 |
4204 |
0.21 |
| chr13_114048059_114048366 | 0.10 |
GRTP1 |
growth hormone regulated TBC protein 1 |
29771 |
0.13 |
| chr4_17782801_17783007 | 0.10 |
FAM184B |
family with sequence similarity 184, member B |
231 |
0.94 |
| chr15_75931462_75932480 | 0.10 |
IMP3 |
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
581 |
0.57 |
| chr9_35907347_35908019 | 0.10 |
HRCT1 |
histidine rich carboxyl terminus 1 |
1494 |
0.3 |
| chr7_100167546_100168141 | 0.10 |
SAP25 |
Sin3A-associated protein, 25kDa |
3427 |
0.11 |
| chr10_123922760_123923208 | 0.10 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
43 |
0.99 |
| chr18_24155058_24155571 | 0.10 |
ENSG00000252846 |
. |
11448 |
0.22 |
| chr8_143695363_143695514 | 0.10 |
ARC |
activity-regulated cytoskeleton-associated protein |
1395 |
0.35 |
| chr13_44947783_44948184 | 0.10 |
SERP2 |
stress-associated endoplasmic reticulum protein family member 2 |
5 |
0.99 |
| chr1_31480017_31480289 | 0.10 |
PUM1 |
pumilio RNA-binding family member 1 |
12202 |
0.19 |
| chr4_124426780_124427488 | 0.10 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
106011 |
0.08 |
| chr9_101469729_101470145 | 0.10 |
GABBR2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
1542 |
0.5 |
| chr9_130330127_130331202 | 0.10 |
FAM129B |
family with sequence similarity 129, member B |
703 |
0.67 |
| chr2_242742783_242743231 | 0.10 |
AC114730.5 |
|
1685 |
0.21 |
| chr12_48513521_48513903 | 0.10 |
PFKM |
phosphofructokinase, muscle |
142 |
0.94 |
| chr9_111205795_111206154 | 0.10 |
ENSG00000222512 |
. |
84765 |
0.11 |
| chr2_5831543_5831945 | 0.09 |
AC107057.2 |
|
124 |
0.83 |
| chr5_180669462_180669613 | 0.09 |
GNB2L1 |
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
291 |
0.58 |
| chr16_65154751_65155029 | 0.09 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
943 |
0.75 |
| chrX_12156590_12157176 | 0.09 |
FRMPD4 |
FERM and PDZ domain containing 4 |
298 |
0.9 |
| chr18_22930019_22930221 | 0.09 |
ZNF521 |
zinc finger protein 521 |
1037 |
0.66 |
| chr2_242157667_242157840 | 0.09 |
ANO7 |
anoctamin 7 |
723 |
0.57 |
| chr9_136857518_136857728 | 0.09 |
VAV2 |
vav 2 guanine nucleotide exchange factor |
103 |
0.97 |
| chr14_72231475_72231648 | 0.09 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
146088 |
0.04 |
| chr19_3367591_3367973 | 0.09 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
1198 |
0.48 |
| chr6_28456937_28457274 | 0.09 |
GPX6 |
glutathione peroxidase 6 (olfactory) |
26459 |
0.14 |
| chr15_44486029_44486998 | 0.09 |
FRMD5 |
FERM domain containing 5 |
131 |
0.98 |
| chr5_42424322_42424771 | 0.09 |
GHR |
growth hormone receptor |
520 |
0.88 |
| chr2_220283280_220283537 | 0.09 |
DES |
desmin |
309 |
0.8 |
| chr16_2835054_2835205 | 0.09 |
PRSS33 |
protease, serine, 33 |
1477 |
0.19 |
| chr5_11384809_11385052 | 0.09 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
203993 |
0.03 |
| chr13_53174146_53174487 | 0.09 |
HNRNPA1L2 |
heterogeneous nuclear ribonucleoprotein A1-like 2 |
17289 |
0.22 |
| chr6_85483496_85483916 | 0.09 |
TBX18 |
T-box 18 |
9469 |
0.3 |
| chr12_3069053_3070004 | 0.09 |
TEAD4 |
TEA domain family member 4 |
445 |
0.77 |
| chr1_159924191_159924342 | 0.09 |
SLAMF9 |
SLAM family member 9 |
222 |
0.88 |
| chr10_123357016_123357770 | 0.09 |
FGFR2 |
fibroblast growth factor receptor 2 |
205 |
0.97 |
| chr1_55436937_55437088 | 0.09 |
TMEM61 |
transmembrane protein 61 |
9453 |
0.16 |
| chr11_11642193_11642674 | 0.09 |
GALNT18 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18 |
1119 |
0.56 |
| chr3_42922021_42922670 | 0.09 |
CYP8B1 |
cytochrome P450, family 8, subfamily B, polypeptide 1 |
4712 |
0.15 |
| chrX_48693575_48693797 | 0.09 |
PCSK1N |
proprotein convertase subtilisin/kexin type 1 inhibitor |
349 |
0.78 |
| chr18_31802690_31803001 | 0.09 |
RP11-379L18.1 |
|
84 |
0.8 |
| chr13_97646306_97646561 | 0.09 |
OXGR1 |
oxoglutarate (alpha-ketoglutarate) receptor 1 |
171 |
0.96 |
| chr7_128045105_128045914 | 0.09 |
IMPDH1 |
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
487 |
0.75 |
| chr3_44040787_44041403 | 0.09 |
ENSG00000252980 |
. |
71484 |
0.12 |
| chr19_36036432_36037158 | 0.09 |
AD000090.2 |
|
136 |
0.59 |
| chr10_81741543_81742003 | 0.09 |
SFTPD |
surfactant protein D |
597 |
0.81 |
| chr1_41961804_41962136 | 0.09 |
EDN2 |
endothelin 2 |
11628 |
0.2 |
| chr2_73518392_73518872 | 0.09 |
EGR4 |
early growth response 4 |
2197 |
0.23 |
| chr5_9547808_9548146 | 0.09 |
ENSG00000239112 |
. |
971 |
0.49 |
| chr5_6449023_6449402 | 0.09 |
UBE2QL1 |
ubiquitin-conjugating enzyme E2Q family-like 1 |
476 |
0.88 |
| chr2_239756978_239757253 | 0.09 |
TWIST2 |
twist family bHLH transcription factor 2 |
442 |
0.88 |
| chr4_54966297_54966506 | 0.09 |
GSX2 |
GS homeobox 2 |
61 |
0.97 |
| chr1_230222759_230222910 | 0.08 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
19816 |
0.23 |
| chr1_8074798_8074959 | 0.08 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
815 |
0.65 |
| chr13_93879365_93880233 | 0.08 |
GPC6 |
glypican 6 |
704 |
0.82 |
| chr13_27936373_27936791 | 0.08 |
ENSG00000252247 |
. |
18699 |
0.15 |
| chr6_1611333_1611591 | 0.08 |
FOXC1 |
forkhead box C1 |
781 |
0.77 |
| chr2_240169087_240169238 | 0.08 |
HDAC4 |
histone deacetylase 4 |
56410 |
0.1 |
| chr3_44803922_44804142 | 0.08 |
KIF15 |
kinesin family member 15 |
678 |
0.45 |
| chr3_48722741_48723287 | 0.08 |
NCKIPSD |
NCK interacting protein with SH3 domain |
254 |
0.86 |
| chr16_48570079_48570232 | 0.08 |
RP11-44I10.3 |
|
23417 |
0.17 |
| chr1_92495646_92496322 | 0.08 |
EPHX4 |
epoxide hydrolase 4 |
445 |
0.85 |
| chr11_70244216_70245100 | 0.08 |
CTTN |
cortactin |
11 |
0.53 |
| chr6_28574388_28574671 | 0.08 |
SCAND3 |
SCAN domain containing 3 |
19417 |
0.18 |
| chr6_84139529_84139855 | 0.08 |
ME1 |
malic enzyme 1, NADP(+)-dependent, cytosolic |
1072 |
0.62 |
| chr4_157862974_157863267 | 0.08 |
PDGFC |
platelet derived growth factor C |
28935 |
0.19 |
| chr5_150325496_150325780 | 0.08 |
ZNF300P1 |
zinc finger protein 300 pseudogene 1 |
213 |
0.94 |
| chr17_40932934_40933198 | 0.08 |
WNK4 |
WNK lysine deficient protein kinase 4 |
370 |
0.71 |
| chr19_17422589_17422876 | 0.08 |
ABHD8 |
abhydrolase domain containing 8 |
1687 |
0.15 |
| chrX_114467670_114468385 | 0.08 |
LRCH2 |
leucine-rich repeats and calponin homology (CH) domain containing 2 |
601 |
0.77 |
| chr9_116918690_116918954 | 0.08 |
COL27A1 |
collagen, type XXVII, alpha 1 |
982 |
0.6 |
| chr18_70534206_70534360 | 0.08 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
17 |
0.98 |
| chr21_35348616_35348984 | 0.08 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
6980 |
0.21 |
| chr19_16179581_16179939 | 0.08 |
TPM4 |
tropomyosin 4 |
1250 |
0.44 |
| chr17_8533666_8534137 | 0.08 |
MYH10 |
myosin, heavy chain 10, non-muscle |
134 |
0.97 |
| chr13_21295723_21296224 | 0.08 |
ENSG00000265710 |
. |
18131 |
0.18 |
| chr19_55658159_55658310 | 0.08 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
76 |
0.93 |
| chr14_105965982_105966133 | 0.08 |
C14orf80 |
chromosome 14 open reading frame 80 |
8326 |
0.11 |
| chr8_23711375_23711668 | 0.08 |
STC1 |
stanniocalcin 1 |
303 |
0.92 |
| chr10_134581493_134581644 | 0.08 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
2240 |
0.34 |
| chr4_154149984_154150203 | 0.08 |
TRIM2 |
tripartite motif containing 2 |
24475 |
0.17 |
| chr6_160769471_160769667 | 0.08 |
SLC22A3 |
solute carrier family 22 (organic cation transporter), member 3 |
144 |
0.98 |
| chr5_15500636_15500842 | 0.08 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
434 |
0.91 |
| chr20_9048816_9049419 | 0.08 |
PLCB4 |
phospholipase C, beta 4 |
293 |
0.9 |
| chr21_35319646_35320126 | 0.08 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
1144 |
0.46 |
| chr5_343986_344609 | 0.08 |
AHRR |
aryl-hydrocarbon receptor repressor |
654 |
0.71 |
| chr11_65666966_65667198 | 0.08 |
FOSL1 |
FOS-like antigen 1 |
808 |
0.38 |
| chr22_39638932_39639142 | 0.08 |
PDGFB |
platelet-derived growth factor beta polypeptide |
16 |
0.97 |
| chr15_63664855_63665177 | 0.08 |
CA12 |
carbonic anhydrase XII |
9018 |
0.26 |
| chr16_447351_447528 | 0.08 |
NME4 |
NME/NM23 nucleoside diphosphate kinase 4 |
202 |
0.88 |
| chr3_43935752_43936459 | 0.08 |
ENSG00000252980 |
. |
176474 |
0.03 |
| chr2_127643626_127643777 | 0.08 |
AC114783.1 |
Protein LOC339760 |
12771 |
0.28 |
| chr9_35116014_35116425 | 0.08 |
FAM214B |
family with sequence similarity 214, member B |
119 |
0.94 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 | 0.0 | GO:0046084 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.1 | GO:0010667 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage development(GO:0060534) trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.0 | GO:0060430 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.0 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.0 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.0 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.0 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID IGF1 PATHWAY | IGF1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |