Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
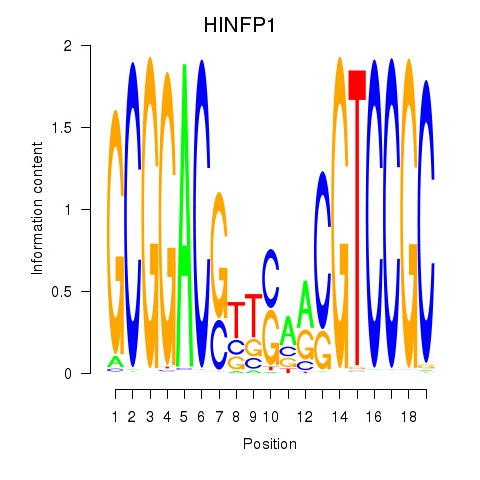
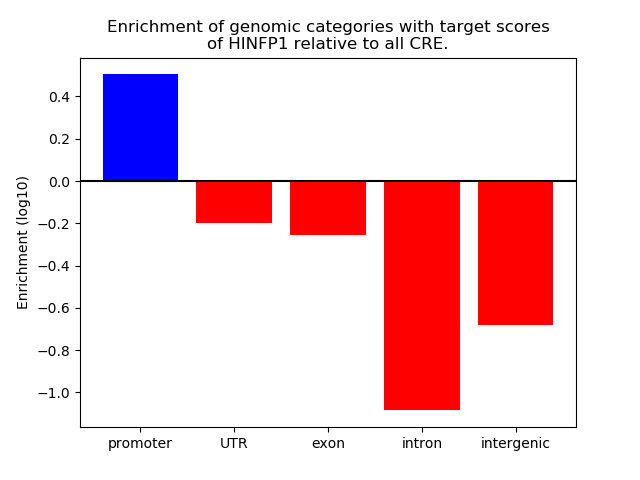
Results for HINFP1
Z-value: 0.56
Transcription factors associated with HINFP1
| Gene Symbol | Gene ID | Gene Info |
|---|
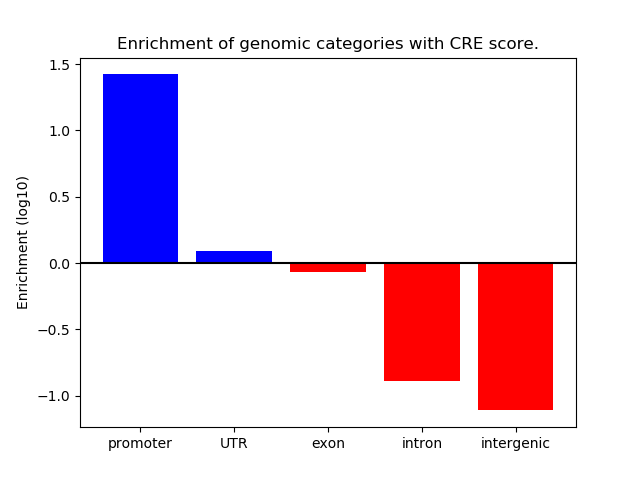
Activity of the HINFP1 motif across conditions
Conditions sorted by the z-value of the HINFP1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
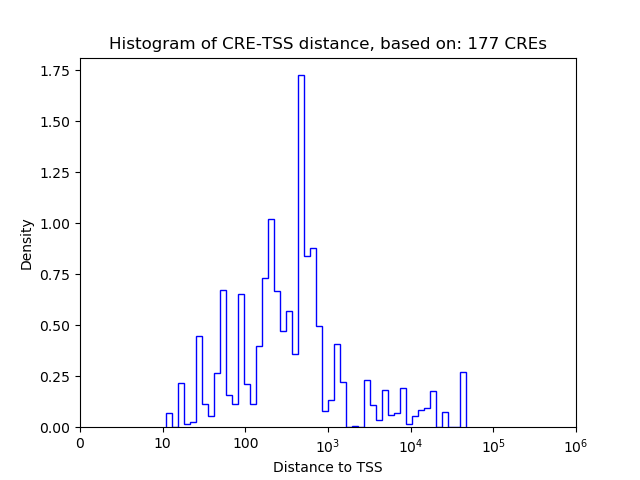
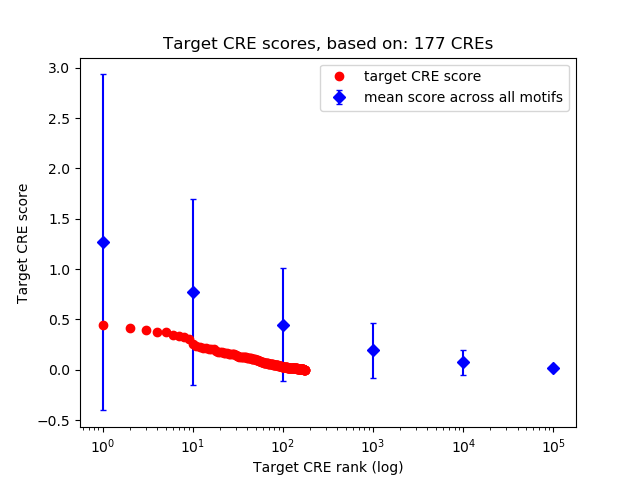
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_30454378_30455881 | 0.45 |
LBH |
limb bud and heart development |
83 |
0.98 |
| chr20_3994903_3996170 | 0.42 |
RNF24 |
ring finger protein 24 |
500 |
0.83 |
| chr17_57408824_57410302 | 0.40 |
YPEL2 |
yippee-like 2 (Drosophila) |
500 |
0.78 |
| chr1_62784626_62784933 | 0.38 |
KANK4 |
KN motif and ankyrin repeat domains 4 |
193 |
0.95 |
| chr2_96809842_96811219 | 0.38 |
DUSP2 |
dual specificity phosphatase 2 |
649 |
0.65 |
| chr3_96531977_96533205 | 0.34 |
EPHA6 |
EPH receptor A6 |
834 |
0.76 |
| chr7_26415747_26416790 | 0.34 |
AC004540.4 |
|
53 |
0.98 |
| chr1_169454193_169456202 | 0.32 |
SLC19A2 |
solute carrier family 19 (thiamine transporter), member 2 |
28 |
0.98 |
| chr16_67197260_67198336 | 0.31 |
HSF4 |
heat shock transcription factor 4 |
510 |
0.52 |
| chr1_231297648_231298418 | 0.26 |
TRIM67 |
tripartite motif containing 67 |
175 |
0.95 |
| chr20_708643_709189 | 0.24 |
SLC52A3 |
solute carrier family 52 (riboflavin transporter), member 3 |
40215 |
0.13 |
| chr19_2251617_2252237 | 0.23 |
ENSG00000267021 |
. |
1289 |
0.21 |
| chr1_11724348_11725085 | 0.21 |
FBXO6 |
F-box protein 6 |
535 |
0.67 |
| chr18_47017567_47018883 | 0.21 |
RPL17 |
ribosomal protein L17 |
15 |
0.51 |
| chr20_35974122_35974616 | 0.20 |
SRC |
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
188 |
0.96 |
| chr17_79875057_79876036 | 0.20 |
SIRT7 |
sirtuin 7 |
524 |
0.46 |
| chr10_62492315_62493446 | 0.20 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
368 |
0.92 |
| chr11_120894558_120895785 | 0.19 |
TBCEL |
tubulin folding cofactor E-like |
341 |
0.93 |
| chr13_22032188_22033345 | 0.18 |
ZDHHC20 |
zinc finger, DHHC-type containing 20 |
648 |
0.76 |
| chr22_24989005_24989426 | 0.18 |
FAM211B |
family with sequence similarity 211, member B |
185 |
0.9 |
| chr3_196695819_196696888 | 0.17 |
PIGZ |
phosphatidylinositol glycan anchor biosynthesis, class Z |
611 |
0.66 |
| chr8_69242814_69243988 | 0.17 |
C8orf34 |
chromosome 8 open reading frame 34 |
56 |
0.82 |
| chr7_101460813_101461304 | 0.17 |
CUX1 |
cut-like homeobox 1 |
138 |
0.97 |
| chr6_163833862_163834944 | 0.17 |
QKI |
QKI, KH domain containing, RNA binding |
1272 |
0.63 |
| chr13_20766378_20767168 | 0.16 |
GJB2 |
gap junction protein, beta 2, 26kDa |
264 |
0.91 |
| chr12_90101448_90102652 | 0.15 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
558 |
0.78 |
| chr3_160822448_160823114 | 0.15 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
43 |
0.67 |
| chr4_187025490_187026623 | 0.15 |
FAM149A |
family with sequence similarity 149, member A |
175 |
0.94 |
| chr18_12308279_12308732 | 0.15 |
TUBB6 |
tubulin, beta 6 class V |
243 |
0.92 |
| chr1_935525_936213 | 0.15 |
HES4 |
hes family bHLH transcription factor 4 |
317 |
0.77 |
| chr1_236958581_236959780 | 0.14 |
MTR |
5-methyltetrahydrofolate-homocysteine methyltransferase |
198 |
0.96 |
| chr10_123733982_123734880 | 0.14 |
NSMCE4A |
non-SMC element 4 homolog A (S. cerevisiae) |
158 |
0.96 |
| chr2_71693301_71694189 | 0.13 |
DYSF |
dysferlin |
87 |
0.98 |
| chr9_86595749_86596582 | 0.13 |
RMI1 |
RecQ mediated genome instability 1 |
452 |
0.51 |
| chr16_84733607_84734797 | 0.13 |
USP10 |
ubiquitin specific peptidase 10 |
556 |
0.79 |
| chr12_95942120_95942537 | 0.13 |
USP44 |
ubiquitin specific peptidase 44 |
246 |
0.94 |
| chr9_131418722_131419532 | 0.13 |
WDR34 |
WD repeat domain 34 |
61 |
0.96 |
| chr7_111201649_111202512 | 0.13 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
100 |
0.98 |
| chr19_13056666_13057332 | 0.13 |
RAD23A |
RAD23 homolog A (S. cerevisiae) |
293 |
0.71 |
| chr20_1875023_1875335 | 0.12 |
SIRPA |
signal-regulatory protein alpha |
25 |
0.98 |
| chrX_70150677_70150828 | 0.12 |
SLC7A3 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
194 |
0.95 |
| chr6_100061682_100062402 | 0.12 |
PRDM13 |
PR domain containing 13 |
7436 |
0.19 |
| chr10_50606076_50606780 | 0.11 |
DRGX |
dorsal root ganglia homeobox |
2931 |
0.27 |
| chr6_1605443_1605730 | 0.11 |
FOXC1 |
forkhead box C1 |
5095 |
0.33 |
| chr9_37034211_37034515 | 0.11 |
PAX5 |
paired box 5 |
260 |
0.93 |
| chr13_67804057_67805835 | 0.11 |
PCDH9 |
protocadherin 9 |
478 |
0.9 |
| chr1_110753494_110754182 | 0.11 |
KCNC4 |
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
257 |
0.84 |
| chr16_30216869_30217924 | 0.11 |
RP11-347C12.3 |
Uncharacterized protein |
31 |
0.94 |
| chr20_57465496_57466112 | 0.10 |
GNAS |
GNAS complex locus |
553 |
0.66 |
| chr18_3262315_3263346 | 0.10 |
MYL12B |
myosin, light chain 12B, regulatory |
124 |
0.92 |
| chr2_129079627_129080191 | 0.10 |
HS6ST1 |
heparan sulfate 6-O-sulfotransferase 1 |
3758 |
0.29 |
| chr10_94822298_94822949 | 0.10 |
CYP26C1 |
cytochrome P450, family 26, subfamily C, polypeptide 1 |
1602 |
0.34 |
| chr7_149156320_149157211 | 0.10 |
ZNF777 |
zinc finger protein 777 |
1449 |
0.45 |
| chr16_718193_719247 | 0.09 |
RHOT2 |
ras homolog family member T2 |
511 |
0.45 |
| chr5_131745958_131747299 | 0.09 |
C5orf56 |
chromosome 5 open reading frame 56 |
45 |
0.96 |
| chr3_12910219_12910517 | 0.09 |
AC034198.7 |
|
18210 |
0.14 |
| chr8_37620085_37620952 | 0.08 |
PROSC |
proline synthetase co-transcribed homolog (bacterial) |
400 |
0.79 |
| chr6_108497513_108497836 | 0.07 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
7707 |
0.18 |
| chr2_71453255_71454205 | 0.07 |
PAIP2B |
poly(A) binding protein interacting protein 2B |
483 |
0.83 |
| chr6_168720263_168720414 | 0.07 |
DACT2 |
dishevelled-binding antagonist of beta-catenin 2 |
52 |
0.98 |
| chr17_27892819_27893926 | 0.07 |
ABHD15 |
abhydrolase domain containing 15 |
783 |
0.31 |
| chr7_139875207_139876022 | 0.07 |
KDM7A |
lysine (K)-specific demethylase 7A |
1127 |
0.39 |
| chr17_40202025_40202687 | 0.07 |
CTD-2132N18.2 |
|
7221 |
0.11 |
| chr7_100081068_100081412 | 0.07 |
ENSG00000266372 |
. |
226 |
0.55 |
| chr7_92218956_92219793 | 0.07 |
FAM133B |
family with sequence similarity 133, member B |
10 |
0.98 |
| chr6_30457259_30457905 | 0.07 |
HLA-E |
major histocompatibility complex, class I, E |
338 |
0.84 |
| chr6_45631346_45631892 | 0.06 |
ENSG00000252738 |
. |
17778 |
0.29 |
| chr10_112403107_112403970 | 0.06 |
RBM20 |
RNA binding motif protein 20 |
617 |
0.55 |
| chr20_3827532_3828225 | 0.06 |
MAVS |
mitochondrial antiviral signaling protein |
388 |
0.8 |
| chr1_27687344_27687533 | 0.06 |
MAP3K6 |
mitogen-activated protein kinase kinase kinase 6 |
3004 |
0.16 |
| chr10_21823798_21823949 | 0.06 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
195 |
0.92 |
| chr19_4374282_4374693 | 0.06 |
SH3GL1 |
SH3-domain GRB2-like 1 |
5802 |
0.08 |
| chr7_132260669_132261651 | 0.05 |
PLXNA4 |
plexin A4 |
69 |
0.98 |
| chr4_186130212_186131033 | 0.05 |
KIAA1430 |
KIAA1430 |
36 |
0.92 |
| chr22_20306851_20307537 | 0.05 |
DGCR6L |
DiGeorge syndrome critical region gene 6-like |
338 |
0.74 |
| chr14_56585152_56585591 | 0.05 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
278 |
0.94 |
| chr8_143533593_143534231 | 0.05 |
BAI1 |
brain-specific angiogenesis inhibitor 1 |
3121 |
0.27 |
| chr2_178128520_178129352 | 0.05 |
AC079305.10 |
|
151 |
0.76 |
| chr18_29264680_29264911 | 0.05 |
B4GALT6 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
109 |
0.91 |
| chr15_66585571_66586039 | 0.05 |
DIS3L |
DIS3 mitotic control homolog (S. cerevisiae)-like |
80 |
0.53 |
| chr11_70962576_70962972 | 0.05 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
849 |
0.72 |
| chr4_87812935_87814058 | 0.05 |
C4orf36 |
chromosome 4 open reading frame 36 |
52 |
0.98 |
| chr6_29894399_29894823 | 0.05 |
HLA-A |
major histocompatibility complex, class I, A |
14426 |
0.18 |
| chr6_11043780_11044551 | 0.05 |
ELOVL2-AS1 |
ELOVL2 antisense RNA 1 |
172 |
0.73 |
| chr1_156829720_156830506 | 0.05 |
NTRK1 |
neurotrophic tyrosine kinase, receptor, type 1 |
494 |
0.64 |
| chr17_11144173_11144324 | 0.05 |
SHISA6 |
shisa family member 6 |
332 |
0.91 |
| chr1_233463556_233464041 | 0.05 |
MLK4 |
Mitogen-activated protein kinase kinase kinase MLK4 |
284 |
0.94 |
| chr18_11981851_11982483 | 0.04 |
IMPA2 |
inositol(myo)-1(or 4)-monophosphatase 2 |
498 |
0.77 |
| chrY_21154472_21155007 | 0.04 |
ENSG00000252766 |
. |
26234 |
0.25 |
| chr2_136289143_136290038 | 0.04 |
R3HDM1 |
R3H domain containing 1 |
503 |
0.62 |
| chr20_52209104_52210193 | 0.04 |
ZNF217 |
zinc finger protein 217 |
730 |
0.68 |
| chr17_77817761_77818338 | 0.04 |
CBX4 |
chromobox homolog 4 |
4821 |
0.18 |
| chr17_16487582_16488116 | 0.04 |
ZNF287 |
zinc finger protein 287 |
15329 |
0.16 |
| chr1_207082522_207083417 | 0.04 |
IL24 |
interleukin 24 |
11791 |
0.14 |
| chr6_84569350_84570131 | 0.03 |
CYB5R4 |
cytochrome b5 reductase 4 |
365 |
0.9 |
| chr19_50354346_50354967 | 0.03 |
PTOV1 |
prostate tumor overexpressed 1 |
174 |
0.58 |
| chr17_40829888_40830324 | 0.03 |
PLEKHH3 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
1058 |
0.27 |
| chr1_29448339_29449190 | 0.03 |
TMEM200B |
transmembrane protein 200B |
249 |
0.93 |
| chr11_450302_450786 | 0.03 |
PTDSS2 |
phosphatidylserine synthase 2 |
264 |
0.82 |
| chr7_107220684_107221346 | 0.03 |
BCAP29 |
B-cell receptor-associated protein 29 |
102 |
0.86 |
| chr7_96635926_96636202 | 0.03 |
DLX6 |
distal-less homeobox 6 |
673 |
0.48 |
| chr3_14644257_14644427 | 0.03 |
AC090952.5 |
|
46597 |
0.13 |
| chr4_109093413_109093782 | 0.03 |
ENSG00000232021 |
. |
258 |
0.93 |
| chr16_58059170_58059939 | 0.03 |
MMP15 |
matrix metallopeptidase 15 (membrane-inserted) |
84 |
0.96 |
| chr2_33171552_33172146 | 0.03 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
190 |
0.93 |
| chr3_182896879_182897541 | 0.03 |
LAMP3 |
lysosomal-associated membrane protein 3 |
15583 |
0.21 |
| chrX_49033425_49033576 | 0.03 |
PLP2 |
proteolipid protein 2 (colonic epithelium-enriched) |
5227 |
0.09 |
| chr18_32956679_32957498 | 0.03 |
ZNF396 |
zinc finger protein 396 |
186 |
0.96 |
| chr2_232526625_232527001 | 0.03 |
ENSG00000239202 |
. |
15829 |
0.16 |
| chr20_32077934_32078551 | 0.02 |
CBFA2T2 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
319 |
0.89 |
| chr2_234263629_234264304 | 0.02 |
DGKD |
diacylglycerol kinase, delta 130kDa |
812 |
0.51 |
| chr12_58129779_58130334 | 0.02 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
1563 |
0.16 |
| chr6_159360525_159360963 | 0.02 |
ENSG00000223191 |
. |
13141 |
0.19 |
| chr10_8091983_8093018 | 0.02 |
GATA3 |
GATA binding protein 3 |
4156 |
0.36 |
| chr5_159546809_159547203 | 0.02 |
PWWP2A |
PWWP domain containing 2A |
576 |
0.75 |
| chr3_52016673_52017699 | 0.02 |
ABHD14B |
abhydrolase domain containing 14B |
239 |
0.49 |
| chr17_17603690_17604249 | 0.02 |
RAI1 |
retinoic acid induced 1 |
18160 |
0.17 |
| chr5_174871055_174871780 | 0.02 |
DRD1 |
dopamine receptor D1 |
206 |
0.95 |
| chr5_92906290_92906587 | 0.02 |
ENSG00000237187 |
. |
61 |
0.98 |
| chr12_133532729_133533161 | 0.02 |
ZNF605 |
zinc finger protein 605 |
53 |
0.49 |
| chr1_10458416_10458839 | 0.02 |
PGD |
phosphogluconate dehydrogenase |
22 |
0.96 |
| chr3_139654401_139654885 | 0.02 |
CLSTN2 |
calsyntenin 2 |
616 |
0.84 |
| chr11_805194_805716 | 0.02 |
PIDD |
p53-induced death domain protein |
210 |
0.8 |
| chr19_12780320_12781072 | 0.02 |
WDR83 |
WD repeat domain 83 |
179 |
0.62 |
| chr5_139028499_139029117 | 0.02 |
CXXC5 |
CXXC finger protein 5 |
290 |
0.92 |
| chr2_237087502_237088103 | 0.02 |
GBX2 |
gastrulation brain homeobox 2 |
10790 |
0.15 |
| chr9_139439215_139440028 | 0.02 |
NOTCH1 |
notch 1 |
693 |
0.35 |
| chr13_21635697_21635989 | 0.02 |
LATS2 |
large tumor suppressor kinase 2 |
157 |
0.95 |
| chr12_109747933_109748276 | 0.02 |
FOXN4 |
forkhead box N4 |
1079 |
0.55 |
| chr7_32535106_32535501 | 0.02 |
AVL9 |
AVL9 homolog (S. cerevisiase) |
19 |
0.88 |
| chr1_224622519_224623397 | 0.02 |
WDR26 |
WD repeat domain 26 |
957 |
0.59 |
| chr12_27932510_27932994 | 0.02 |
KLHL42 |
kelch-like family member 42 |
201 |
0.87 |
| chr6_24720330_24720995 | 0.02 |
C6orf62 |
chromosome 6 open reading frame 62 |
42 |
0.97 |
| chr13_28528843_28528994 | 0.02 |
ATP5EP2 |
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
9575 |
0.14 |
| chr20_34189521_34189992 | 0.02 |
SPAG4 |
sperm associated antigen 4 |
14058 |
0.11 |
| chr7_75911448_75912023 | 0.01 |
SRRM3 |
serine/arginine repetitive matrix 3 |
197 |
0.93 |
| chr14_45366753_45367297 | 0.01 |
C14orf28 |
chromosome 14 open reading frame 28 |
487 |
0.59 |
| chr1_148176495_148177244 | 0.01 |
PPIAL4D |
peptidylprolyl isomerase A (cyclophilin A)-like 4D |
25667 |
0.21 |
| chr5_6714492_6714756 | 0.01 |
PAPD7 |
PAP associated domain containing 7 |
94 |
0.98 |
| chr11_67275372_67275646 | 0.01 |
CDK2AP2 |
cyclin-dependent kinase 2 associated protein 2 |
53 |
0.93 |
| chr12_19283254_19284293 | 0.01 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
1043 |
0.58 |
| chr5_129239637_129240094 | 0.01 |
CHSY3 |
chondroitin sulfate synthase 3 |
300 |
0.88 |
| chr11_64901308_64901962 | 0.01 |
SYVN1 |
synovial apoptosis inhibitor 1, synoviolin |
265 |
0.76 |
| chr16_75033808_75034560 | 0.01 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
941 |
0.6 |
| chr7_27179265_27179416 | 0.01 |
HOXA3 |
homeobox A3 |
485 |
0.41 |
| chrX_129299242_129299778 | 0.01 |
AIFM1 |
apoptosis-inducing factor, mitochondrion-associated, 1 |
128 |
0.96 |
| chr19_6767133_6767491 | 0.01 |
SH2D3A |
SH2 domain containing 3A |
134 |
0.93 |
| chr6_18277489_18278102 | 0.01 |
ENSG00000199715 |
. |
4298 |
0.21 |
| chr12_6833255_6834039 | 0.01 |
COPS7A |
COP9 signalosome subunit 7A |
175 |
0.85 |
| chr4_190936109_190936296 | 0.01 |
ENSG00000199572 |
. |
91 |
0.95 |
| chr1_24829331_24829634 | 0.01 |
RCAN3 |
RCAN family member 3 |
95 |
0.97 |
| chr7_157072082_157072490 | 0.01 |
ENSG00000266453 |
. |
26201 |
0.21 |
| chr17_46674549_46675551 | 0.01 |
HOXB-AS3 |
HOXB cluster antisense RNA 3 |
1249 |
0.2 |
| chr2_220416827_220417484 | 0.01 |
ENSG00000265252 |
. |
3286 |
0.09 |
| chr17_56234195_56234709 | 0.01 |
OR4D1 |
olfactory receptor, family 4, subfamily D, member 1 |
1958 |
0.26 |
| chr10_119135118_119135768 | 0.01 |
PDZD8 |
PDZ domain containing 8 |
465 |
0.84 |
| chr11_68228271_68228585 | 0.01 |
PPP6R3 |
protein phosphatase 6, regulatory subunit 3 |
147 |
0.96 |
| chr17_12920409_12921278 | 0.01 |
ELAC2 |
elaC ribonuclease Z 2 |
64 |
0.97 |
| chr11_119454980_119455308 | 0.01 |
RP11-196E1.3 |
|
23991 |
0.19 |
| chr17_38296734_38297043 | 0.01 |
CASC3 |
cancer susceptibility candidate 3 |
207 |
0.9 |
| chr5_5421590_5422497 | 0.01 |
KIAA0947 |
KIAA0947 |
764 |
0.8 |
| chr3_141120790_141121159 | 0.01 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
217 |
0.95 |
| chr1_165796835_165797193 | 0.00 |
TMCO1 |
transmembrane and coiled-coil domains 1 |
22 |
0.51 |
| chr19_4304444_4304595 | 0.00 |
FSD1 |
fibronectin type III and SPRY domain containing 1 |
78 |
0.93 |
| chr7_74306205_74306628 | 0.00 |
STAG3L2 |
stromal antigen 3-like 2 (pseudogene) |
259 |
0.91 |
| chr19_16999844_17000017 | 0.00 |
F2RL3 |
coagulation factor II (thrombin) receptor-like 3 |
71 |
0.95 |
| chr7_26192093_26192429 | 0.00 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
401 |
0.85 |
| chr3_50358230_50358862 | 0.00 |
HYAL2 |
hyaluronoglucosaminidase 2 |
34 |
0.93 |
| chr16_77224093_77224603 | 0.00 |
MON1B |
MON1 secretory trafficking family member B |
384 |
0.86 |
| chr6_157801166_157801713 | 0.00 |
ZDHHC14 |
zinc finger, DHHC-type containing 14 |
726 |
0.76 |
| chr16_30662970_30663539 | 0.00 |
PRR14 |
proline rich 14 |
743 |
0.43 |
| chr8_56987192_56987600 | 0.00 |
CTA-397H3.3 |
|
245 |
0.54 |
| chr12_120632090_120632489 | 0.00 |
GCN1L1 |
GCN1 general control of amino-acid synthesis 1-like 1 (yeast) |
224 |
0.88 |
| chr7_89783488_89783754 | 0.00 |
STEAP1 |
six transmembrane epithelial antigen of the prostate 1 |
68 |
0.97 |
| chr22_18893815_18894459 | 0.00 |
DGCR6 |
DiGeorge syndrome critical region gene 6 |
271 |
0.91 |
| chr1_43232974_43233556 | 0.00 |
C1orf50 |
chromosome 1 open reading frame 50 |
288 |
0.65 |
| chr14_23526484_23526858 | 0.00 |
CDH24 |
cadherin 24, type 2 |
76 |
0.93 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.2 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.0 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |