Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
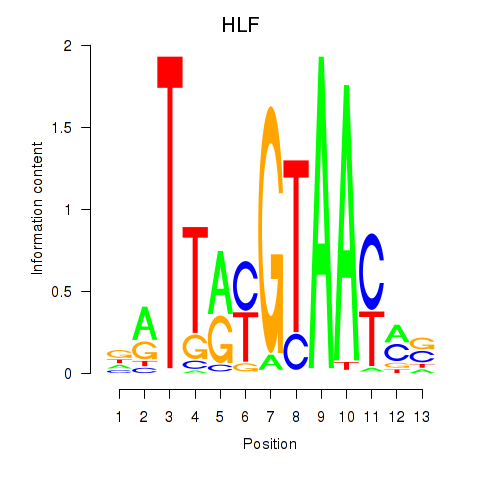
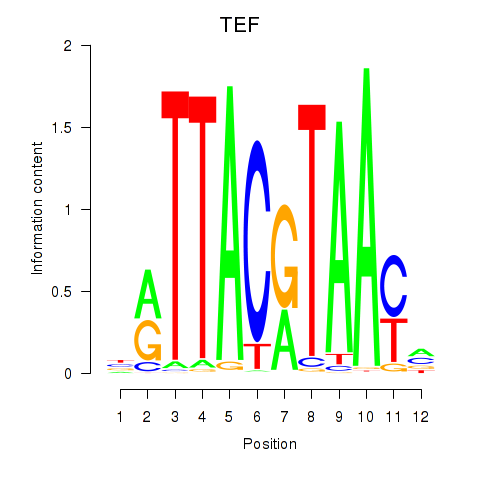
Results for HLF_TEF
Z-value: 0.42
Transcription factors associated with HLF_TEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HLF
|
ENSG00000108924.9 | HLF transcription factor, PAR bZIP family member |
|
TEF
|
ENSG00000167074.10 | TEF transcription factor, PAR bZIP family member |
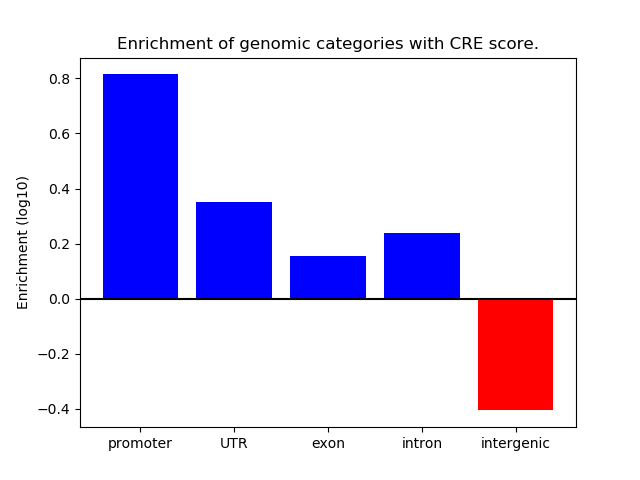
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_53314641_53314792 | HLF | 27657 | 0.210552 | -0.84 | 4.9e-03 | Click! |
| chr17_53276782_53276933 | HLF | 65516 | 0.126106 | -0.75 | 2.1e-02 | Click! |
| chr17_53275978_53276129 | HLF | 66320 | 0.124582 | -0.67 | 5.0e-02 | Click! |
| chr17_53254979_53255130 | HLF | 87319 | 0.090800 | -0.64 | 6.5e-02 | Click! |
| chr17_53341174_53341325 | HLF | 1124 | 0.579188 | -0.45 | 2.2e-01 | Click! |
| chr22_41762903_41763435 | TEF | 168 | 0.935667 | 0.56 | 1.2e-01 | Click! |
| chr22_41777572_41777770 | TEF | 262 | 0.889104 | 0.46 | 2.1e-01 | Click! |
| chr22_41777854_41778145 | TEF | 66 | 0.964245 | 0.33 | 3.9e-01 | Click! |
| chr22_41778564_41778775 | TEF | 270 | 0.885972 | -0.29 | 4.5e-01 | Click! |
| chr22_41800074_41800274 | TEF | 21235 | 0.124993 | 0.27 | 4.8e-01 | Click! |
Activity of the HLF_TEF motif across conditions
Conditions sorted by the z-value of the HLF_TEF motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
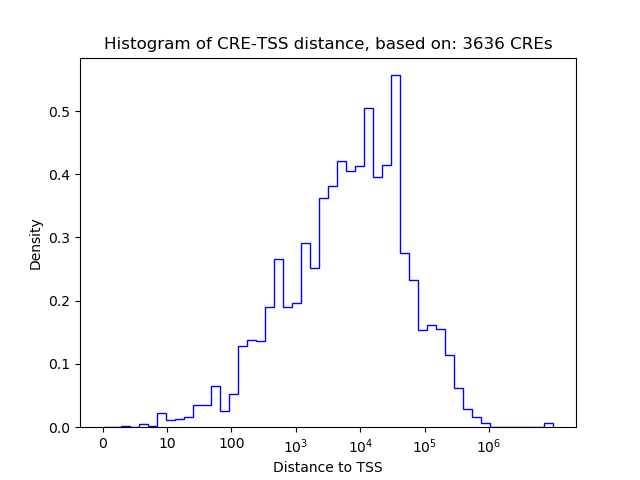
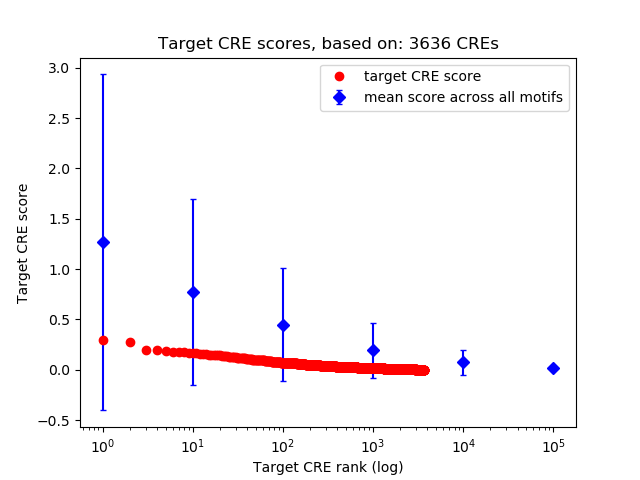
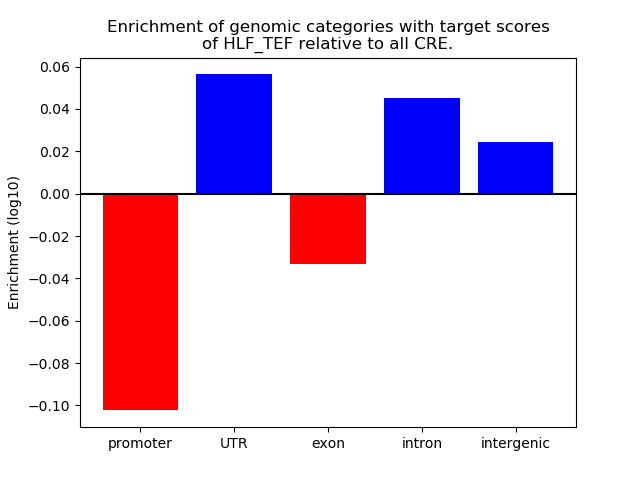
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_36686005_36686156 | 0.29 |
CTD-2353F22.1 |
|
39195 |
0.2 |
| chr3_172280513_172281223 | 0.28 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
39571 |
0.16 |
| chr20_10286915_10287750 | 0.20 |
ENSG00000211588 |
. |
55546 |
0.13 |
| chr15_91955379_91955670 | 0.20 |
SV2B |
synaptic vesicle glycoprotein 2B |
186424 |
0.03 |
| chr2_47788453_47788604 | 0.18 |
KCNK12 |
potassium channel, subfamily K, member 12 |
9550 |
0.18 |
| chr4_38787370_38787521 | 0.18 |
TLR10 |
toll-like receptor 10 |
2835 |
0.22 |
| chr21_37572436_37572587 | 0.17 |
ENSG00000265882 |
. |
13625 |
0.14 |
| chr2_235864048_235864199 | 0.17 |
SH3BP4 |
SH3-domain binding protein 4 |
3403 |
0.4 |
| chr1_210412971_210413268 | 0.17 |
SERTAD4-AS1 |
SERTAD4 antisense RNA 1 |
5727 |
0.24 |
| chr1_17758134_17758285 | 0.17 |
RCC2 |
regulator of chromosome condensation 2 |
6848 |
0.16 |
| chr5_94417062_94417734 | 0.16 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
48 |
0.99 |
| chr2_40682644_40682795 | 0.16 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
2141 |
0.48 |
| chr2_70750715_70751050 | 0.16 |
TGFA |
transforming growth factor, alpha |
29740 |
0.17 |
| chr6_53210504_53210655 | 0.16 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
3008 |
0.28 |
| chr6_31799753_31799904 | 0.15 |
C6orf48 |
chromosome 6 open reading frame 48 |
2557 |
0.08 |
| chr2_28619412_28619563 | 0.15 |
RP11-373D23.2 |
|
195 |
0.87 |
| chrX_24498632_24498874 | 0.15 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
15180 |
0.2 |
| chr18_55334184_55334335 | 0.15 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
21641 |
0.15 |
| chr7_50849637_50849949 | 0.15 |
GRB10 |
growth factor receptor-bound protein 10 |
471 |
0.88 |
| chr9_120407172_120407366 | 0.14 |
TLR4 |
toll-like receptor 4 |
59341 |
0.13 |
| chr9_101871702_101872013 | 0.14 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
4425 |
0.25 |
| chr21_43352965_43353181 | 0.14 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
6274 |
0.19 |
| chr3_170078673_170078995 | 0.14 |
SKIL |
SKI-like oncogene |
1423 |
0.5 |
| chr11_64647105_64647256 | 0.14 |
EHD1 |
EH-domain containing 1 |
8 |
0.95 |
| chr2_100107380_100107531 | 0.13 |
REV1 |
REV1, polymerase (DNA directed) |
958 |
0.61 |
| chrX_153966699_153966850 | 0.13 |
GAB3 |
GRB2-associated binding protein 3 |
12558 |
0.13 |
| chr2_163206460_163206611 | 0.13 |
GCA |
grancalcin, EF-hand calcium binding protein |
5687 |
0.26 |
| chr12_11809211_11809577 | 0.13 |
ETV6 |
ets variant 6 |
6606 |
0.29 |
| chr12_93988367_93988869 | 0.13 |
SOCS2 |
suppressor of cytokine signaling 2 |
19784 |
0.17 |
| chr17_65430423_65430574 | 0.13 |
ENSG00000244610 |
. |
22908 |
0.12 |
| chr7_150786138_150786598 | 0.12 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
2214 |
0.15 |
| chr10_125847748_125848017 | 0.12 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
4088 |
0.34 |
| chr4_47912581_47912732 | 0.12 |
NFXL1 |
nuclear transcription factor, X-box binding-like 1 |
3900 |
0.24 |
| chr7_41515963_41516114 | 0.12 |
INHBA-AS1 |
INHBA antisense RNA 1 |
217476 |
0.02 |
| chr18_33164100_33164377 | 0.12 |
GALNT1 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) |
2534 |
0.31 |
| chr11_5307307_5307601 | 0.11 |
OR51B4 |
olfactory receptor, family 51, subfamily B, member 4 |
15772 |
0.08 |
| chr8_17193981_17194132 | 0.11 |
MTMR7 |
myotubularin related protein 7 |
24874 |
0.21 |
| chr7_42965840_42965991 | 0.11 |
PSMA2 |
proteasome (prosome, macropain) subunit, alpha type, 2 |
5858 |
0.2 |
| chr1_234737377_234737528 | 0.11 |
RP4-781K5.2 |
|
5302 |
0.2 |
| chr2_160470399_160470706 | 0.11 |
AC009506.1 |
|
1255 |
0.4 |
| chr11_78101759_78101910 | 0.11 |
RP11-452H21.2 |
|
2727 |
0.27 |
| chr10_112033444_112033726 | 0.11 |
SMNDC1 |
survival motor neuron domain containing 1 |
30870 |
0.18 |
| chr13_48811096_48811587 | 0.11 |
ITM2B |
integral membrane protein 2B |
4002 |
0.31 |
| chrX_49872160_49872311 | 0.10 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
38079 |
0.11 |
| chr20_6749993_6750220 | 0.10 |
BMP2 |
bone morphogenetic protein 2 |
1795 |
0.51 |
| chr10_17269607_17270502 | 0.10 |
VIM |
vimentin |
204 |
0.89 |
| chr11_112027837_112027988 | 0.10 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
6885 |
0.11 |
| chr9_70616019_70616170 | 0.10 |
AL591479.1 |
Uncharacterized protein |
30495 |
0.22 |
| chr10_51574545_51574696 | 0.10 |
NCOA4 |
nuclear receptor coactivator 4 |
1320 |
0.42 |
| chr2_42979179_42979330 | 0.10 |
OXER1 |
oxoeicosanoid (OXE) receptor 1 |
12147 |
0.21 |
| chr3_152884468_152884748 | 0.10 |
RAP2B |
RAP2B, member of RAS oncogene family |
4579 |
0.23 |
| chr6_135061410_135061561 | 0.10 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
188814 |
0.03 |
| chr1_243430959_243431113 | 0.10 |
SDCCAG8 |
serologically defined colon cancer antigen 8 |
11664 |
0.19 |
| chr19_2236436_2236682 | 0.10 |
SF3A2 |
splicing factor 3a, subunit 2, 66kDa |
39 |
0.72 |
| chr2_70145279_70145482 | 0.10 |
MXD1 |
MAX dimerization protein 1 |
3060 |
0.16 |
| chr10_51575835_51576189 | 0.10 |
NCOA4 |
nuclear receptor coactivator 4 |
273 |
0.91 |
| chr5_135470545_135470952 | 0.10 |
SMAD5-AS1 |
SMAD5 antisense RNA 1 |
169 |
0.92 |
| chr9_94184850_94185394 | 0.10 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
1022 |
0.67 |
| chr22_19874685_19874970 | 0.09 |
GNB1L |
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
32365 |
0.11 |
| chr8_23408711_23408862 | 0.09 |
AC051642.1 |
|
11865 |
0.15 |
| chr10_77190951_77191328 | 0.09 |
RP11-399K21.10 |
|
207 |
0.95 |
| chr7_127640727_127640878 | 0.09 |
LRRC4 |
leucine rich repeat containing 4 |
30256 |
0.2 |
| chr22_23775353_23775544 | 0.09 |
ZDHHC8P1 |
zinc finger, DHHC-type containing 8 pseudogene 1 |
31104 |
0.16 |
| chr17_51213191_51213342 | 0.09 |
C17orf112 |
chromosome 17 open reading frame 112 |
150386 |
0.05 |
| chr6_27570423_27570827 | 0.09 |
ENSG00000206671 |
. |
6434 |
0.22 |
| chr17_16171067_16171218 | 0.09 |
ENSG00000221355 |
. |
14186 |
0.16 |
| chr1_200558260_200558411 | 0.09 |
KIF14 |
kinesin family member 14 |
31527 |
0.2 |
| chr2_219093418_219093644 | 0.09 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
11598 |
0.11 |
| chr6_82519484_82519635 | 0.09 |
ENSG00000206886 |
. |
45818 |
0.16 |
| chr19_52255571_52255845 | 0.09 |
FPR2 |
formyl peptide receptor 2 |
429 |
0.51 |
| chr9_140487358_140487509 | 0.09 |
ZMYND19 |
zinc finger, MYND-type containing 19 |
2491 |
0.17 |
| chr2_106416905_106417056 | 0.08 |
NCK2 |
NCK adaptor protein 2 |
16035 |
0.26 |
| chr14_72064432_72064583 | 0.08 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
444 |
0.9 |
| chr8_142246332_142246483 | 0.08 |
SLC45A4 |
solute carrier family 45, member 4 |
5960 |
0.16 |
| chr15_93351259_93351410 | 0.08 |
CTD-2313J17.1 |
|
463 |
0.75 |
| chr1_192139800_192139951 | 0.08 |
RGS18 |
regulator of G-protein signaling 18 |
12288 |
0.31 |
| chr7_151620422_151620573 | 0.08 |
ENSG00000222941 |
. |
7085 |
0.15 |
| chr12_58818401_58819089 | 0.08 |
RP11-362K2.2 |
Protein LOC100506869 |
119162 |
0.06 |
| chr18_13464907_13465379 | 0.08 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
129 |
0.93 |
| chr10_119129819_119130103 | 0.08 |
PDZD8 |
PDZ domain containing 8 |
5017 |
0.24 |
| chr13_92500241_92500392 | 0.08 |
ENSG00000252508 |
. |
211781 |
0.03 |
| chr8_23019732_23020469 | 0.08 |
TNFRSF10D |
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
1443 |
0.33 |
| chr6_134551187_134551338 | 0.08 |
ENSG00000238631 |
. |
23610 |
0.16 |
| chr5_124254833_124254984 | 0.08 |
ZNF608 |
zinc finger protein 608 |
170408 |
0.03 |
| chr1_240073490_240073887 | 0.08 |
CHRM3-AS1 |
CHRM3 antisense RNA 1 |
10516 |
0.29 |
| chr14_73928931_73929214 | 0.08 |
ENSG00000251393 |
. |
57 |
0.97 |
| chr15_52167016_52167539 | 0.08 |
ENSG00000207484 |
. |
9476 |
0.13 |
| chr10_43828186_43828337 | 0.08 |
ENSG00000221468 |
. |
8973 |
0.19 |
| chr9_80572297_80572448 | 0.08 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
73148 |
0.13 |
| chr14_65183552_65183703 | 0.08 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
1206 |
0.53 |
| chr12_65143392_65143687 | 0.07 |
GNS |
glucosamine (N-acetyl)-6-sulfatase |
3112 |
0.19 |
| chr3_120277883_120278034 | 0.07 |
NDUFB4 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
37198 |
0.19 |
| chr16_48645428_48645579 | 0.07 |
N4BP1 |
NEDD4 binding protein 1 |
1383 |
0.44 |
| chr2_207998546_207999225 | 0.07 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
24 |
0.98 |
| chr10_11328636_11328787 | 0.07 |
CELF2-AS1 |
CELF2 antisense RNA 1 |
33136 |
0.21 |
| chr3_165179707_165179858 | 0.07 |
SLITRK3 |
SLIT and NTRK-like family, member 3 |
264885 |
0.02 |
| chr17_40608681_40609216 | 0.07 |
ATP6V0A1 |
ATPase, H+ transporting, lysosomal V0 subunit a1 |
1914 |
0.19 |
| chr11_1320381_1320746 | 0.07 |
TOLLIP |
toll interacting protein |
3457 |
0.18 |
| chr6_137477199_137477350 | 0.07 |
IL22RA2 |
interleukin 22 receptor, alpha 2 |
17511 |
0.24 |
| chr13_108457470_108457621 | 0.07 |
FAM155A-IT1 |
FAM155A intronic transcript 1 (non-protein coding) |
30254 |
0.22 |
| chr2_212989643_212989794 | 0.07 |
ENSG00000221782 |
. |
301366 |
0.01 |
| chr20_47441337_47441845 | 0.07 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
2829 |
0.36 |
| chr2_38143594_38143745 | 0.07 |
RMDN2 |
regulator of microtubule dynamics 2 |
6661 |
0.29 |
| chr1_151430384_151431013 | 0.07 |
POGZ |
pogo transposable element with ZNF domain |
957 |
0.44 |
| chr3_171779467_171780048 | 0.07 |
FNDC3B |
fibronectin type III domain containing 3B |
15054 |
0.27 |
| chr3_150919418_150919700 | 0.07 |
GPR171 |
G protein-coupled receptor 171 |
1420 |
0.35 |
| chr4_13530041_13530301 | 0.07 |
NKX3-2 |
NK3 homeobox 2 |
16503 |
0.21 |
| chr9_21023989_21024197 | 0.07 |
PTPLAD2 |
protein tyrosine phosphatase-like A domain containing 2 |
7515 |
0.22 |
| chr4_100738641_100738957 | 0.07 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
796 |
0.72 |
| chr10_114344566_114344899 | 0.07 |
ENSG00000264763 |
. |
49197 |
0.15 |
| chr22_35805907_35806058 | 0.07 |
MCM5 |
minichromosome maintenance complex component 5 |
9558 |
0.2 |
| chr3_172240144_172241280 | 0.07 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
553 |
0.83 |
| chr4_176867859_176868338 | 0.07 |
GPM6A |
glycoprotein M6A |
24000 |
0.23 |
| chr5_49944181_49944332 | 0.07 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
17477 |
0.3 |
| chr16_9193995_9194146 | 0.07 |
RP11-473I1.9 |
|
4635 |
0.17 |
| chr9_70496557_70496726 | 0.07 |
CBWD5 |
COBW domain containing 5 |
6395 |
0.28 |
| chr3_171470368_171470519 | 0.07 |
PLD1 |
phospholipase D1, phosphatidylcholine-specific |
18706 |
0.21 |
| chr14_101909161_101909312 | 0.07 |
ENSG00000258498 |
. |
117523 |
0.04 |
| chr8_19421258_19421409 | 0.07 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
38043 |
0.23 |
| chr9_94186787_94187440 | 0.07 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
969 |
0.69 |
| chr7_87128175_87128326 | 0.07 |
ABCB4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
18499 |
0.24 |
| chr7_75535458_75535737 | 0.07 |
POR |
P450 (cytochrome) oxidoreductase |
1297 |
0.38 |
| chr14_54914007_54914158 | 0.07 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
5933 |
0.22 |
| chr3_128964457_128964651 | 0.07 |
COPG1 |
coatomer protein complex, subunit gamma 1 |
3895 |
0.19 |
| chr10_3710614_3710914 | 0.07 |
RP11-184A2.3 |
|
82495 |
0.1 |
| chr8_67042351_67042768 | 0.07 |
TRIM55 |
tripartite motif containing 55 |
3281 |
0.32 |
| chr1_207509019_207509170 | 0.07 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
13958 |
0.26 |
| chr4_95489578_95489729 | 0.06 |
PDLIM5 |
PDZ and LIM domain 5 |
44777 |
0.21 |
| chr17_38269362_38269513 | 0.06 |
MSL1 |
male-specific lethal 1 homolog (Drosophila) |
9114 |
0.11 |
| chr6_26304546_26304697 | 0.06 |
HIST1H4H |
histone cluster 1, H4h |
18859 |
0.08 |
| chr11_59950051_59950297 | 0.06 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
325 |
0.9 |
| chr1_206857600_206857751 | 0.06 |
MAPKAPK2 |
mitogen-activated protein kinase-activated protein kinase 2 |
614 |
0.64 |
| chr11_118109635_118109786 | 0.06 |
MPZL3 |
myelin protein zero-like 3 |
13352 |
0.13 |
| chr13_48809687_48810120 | 0.06 |
ITM2B |
integral membrane protein 2B |
2564 |
0.37 |
| chr3_15358347_15358781 | 0.06 |
ENSG00000238891 |
. |
437 |
0.79 |
| chr1_169580216_169580367 | 0.06 |
SELP |
selectin P (granule membrane protein 140kDa, antigen CD62) |
8166 |
0.21 |
| chrX_11434732_11434905 | 0.06 |
ARHGAP6 |
Rho GTPase activating protein 6 |
11075 |
0.29 |
| chr7_36763519_36763817 | 0.06 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
386 |
0.9 |
| chr2_100105997_100106148 | 0.06 |
REV1 |
REV1, polymerase (DNA directed) |
363 |
0.89 |
| chr7_93694115_93694266 | 0.06 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
60496 |
0.13 |
| chr9_97629545_97629745 | 0.06 |
RP11-49O14.2 |
|
33066 |
0.14 |
| chr4_175525550_175525701 | 0.06 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
81320 |
0.11 |
| chr11_77113152_77113303 | 0.06 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
9722 |
0.24 |
| chr11_23250751_23251044 | 0.06 |
ENSG00000264478 |
. |
242362 |
0.02 |
| chr9_70850276_70850542 | 0.06 |
CBWD3 |
COBW domain containing 3 |
5988 |
0.18 |
| chr14_53301814_53302501 | 0.06 |
FERMT2 |
fermitin family member 2 |
29082 |
0.15 |
| chr13_32620855_32621006 | 0.06 |
FRY |
furry homolog (Drosophila) |
14071 |
0.22 |
| chr11_33913285_33913515 | 0.06 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
436 |
0.85 |
| chrX_9431281_9431551 | 0.06 |
TBL1X |
transducin (beta)-like 1X-linked |
45 |
0.99 |
| chr21_34712478_34712629 | 0.06 |
IFNAR1 |
interferon (alpha, beta and omega) receptor 1 |
15275 |
0.17 |
| chrX_27101459_27101705 | 0.06 |
ENSG00000252486 |
. |
168770 |
0.04 |
| chr11_119598721_119599273 | 0.06 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
266 |
0.88 |
| chr17_55970381_55971074 | 0.06 |
CUEDC1 |
CUE domain containing 1 |
678 |
0.68 |
| chr3_186633899_186634165 | 0.06 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
14242 |
0.17 |
| chr1_185014704_185016028 | 0.06 |
RNF2 |
ring finger protein 2 |
138 |
0.97 |
| chr8_56792958_56793527 | 0.06 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
848 |
0.55 |
| chr2_60783649_60784435 | 0.06 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
3340 |
0.31 |
| chr9_92756542_92756693 | 0.06 |
ENSG00000263967 |
. |
29200 |
0.27 |
| chr5_169301696_169301847 | 0.06 |
CTB-37A13.1 |
|
95402 |
0.08 |
| chr8_129262697_129262848 | 0.06 |
ENSG00000201782 |
. |
30022 |
0.23 |
| chr2_68654340_68654491 | 0.06 |
FBXO48 |
F-box protein 48 |
39975 |
0.13 |
| chr2_167231853_167232143 | 0.06 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
499 |
0.84 |
| chr1_68149030_68149645 | 0.06 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
1407 |
0.47 |
| chr1_41326478_41326629 | 0.06 |
CITED4 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
1465 |
0.35 |
| chr2_197025134_197025564 | 0.06 |
STK17B |
serine/threonine kinase 17b |
3978 |
0.23 |
| chr9_135548954_135549215 | 0.06 |
DDX31 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
3296 |
0.24 |
| chr17_74547322_74547606 | 0.06 |
RP11-666A8.12 |
|
30 |
0.69 |
| chr7_151696730_151696881 | 0.06 |
GALNT11 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
25973 |
0.14 |
| chr8_18769114_18769265 | 0.06 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
24645 |
0.22 |
| chr15_89637238_89637389 | 0.06 |
ABHD2 |
abhydrolase domain containing 2 |
5652 |
0.19 |
| chr5_95109290_95109585 | 0.06 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
5593 |
0.19 |
| chr15_101112185_101112336 | 0.05 |
ENSG00000200095 |
. |
11263 |
0.13 |
| chr18_56282753_56282991 | 0.05 |
ALPK2 |
alpha-kinase 2 |
13317 |
0.15 |
| chr16_66555734_66555905 | 0.05 |
TK2 |
thymidine kinase 2, mitochondrial |
8133 |
0.11 |
| chr12_64991986_64992137 | 0.05 |
RP11-338E21.1 |
|
11178 |
0.13 |
| chr4_154582407_154582558 | 0.05 |
RP11-153M7.3 |
|
16929 |
0.19 |
| chr10_111827720_111827871 | 0.05 |
ADD3 |
adducin 3 (gamma) |
60073 |
0.12 |
| chr20_62521666_62521817 | 0.05 |
DNAJC5 |
DnaJ (Hsp40) homolog, subfamily C, member 5 |
4777 |
0.08 |
| chr18_52274836_52275204 | 0.05 |
DYNAP |
dynactin associated protein |
16630 |
0.28 |
| chr20_49026268_49026419 | 0.05 |
ENSG00000244376 |
. |
19677 |
0.22 |
| chr4_187765993_187766245 | 0.05 |
ENSG00000252382 |
. |
12491 |
0.3 |
| chr8_68515024_68515175 | 0.05 |
ENSG00000221660 |
. |
108768 |
0.07 |
| chr12_122884260_122884886 | 0.05 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
12 |
0.98 |
| chr2_190140835_190141232 | 0.05 |
ENSG00000266817 |
. |
35454 |
0.2 |
| chr6_144128825_144128976 | 0.05 |
LTV1 |
LTV1 homolog (S. cerevisiae) |
35581 |
0.19 |
| chr3_94124571_94124722 | 0.05 |
ENSG00000253062 |
. |
307236 |
0.01 |
| chr17_19172425_19172576 | 0.05 |
EPN2 |
epsin 2 |
11267 |
0.12 |
| chr17_76588567_76588821 | 0.05 |
DNAH17 |
dynein, axonemal, heavy chain 17 |
15218 |
0.19 |
| chr16_87905110_87905261 | 0.05 |
SLC7A5 |
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
2091 |
0.29 |
| chr1_154130279_154130430 | 0.05 |
NUP210L |
nucleoporin 210kDa-like |
2762 |
0.15 |
| chr6_27791567_27792629 | 0.05 |
HIST1H4J |
histone cluster 1, H4j |
214 |
0.82 |
| chr2_201542522_201542673 | 0.05 |
AOX1 |
aldehyde oxidase 1 |
14967 |
0.21 |
| chr6_72001840_72001991 | 0.05 |
OGFRL1 |
opioid growth factor receptor-like 1 |
3409 |
0.26 |
| chr17_80398389_80398540 | 0.05 |
HEXDC |
hexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing |
3859 |
0.1 |
| chr3_42132456_42132951 | 0.05 |
TRAK1 |
trafficking protein, kinesin binding 1 |
141 |
0.98 |
| chr15_52436530_52436681 | 0.05 |
CTD-2184D3.7 |
|
5980 |
0.16 |
| chr1_151579139_151579290 | 0.05 |
SNX27 |
sorting nexin family member 27 |
5344 |
0.12 |
| chr10_99223651_99223802 | 0.05 |
MMS19 |
MMS19 nucleotide excision repair homolog (S. cerevisiae) |
3556 |
0.12 |
| chr6_144357186_144358268 | 0.05 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
28008 |
0.21 |
| chr8_30581756_30582064 | 0.05 |
GSR |
glutathione reductase |
2617 |
0.31 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0003171 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.0 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.0 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |