Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
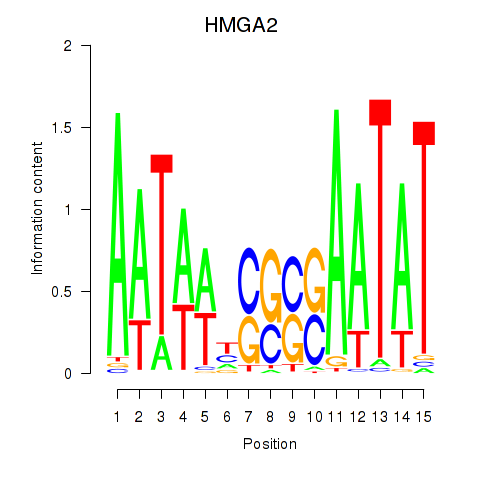
Results for HMGA2
Z-value: 0.56
Transcription factors associated with HMGA2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMGA2
|
ENSG00000149948.9 | high mobility group AT-hook 2 |
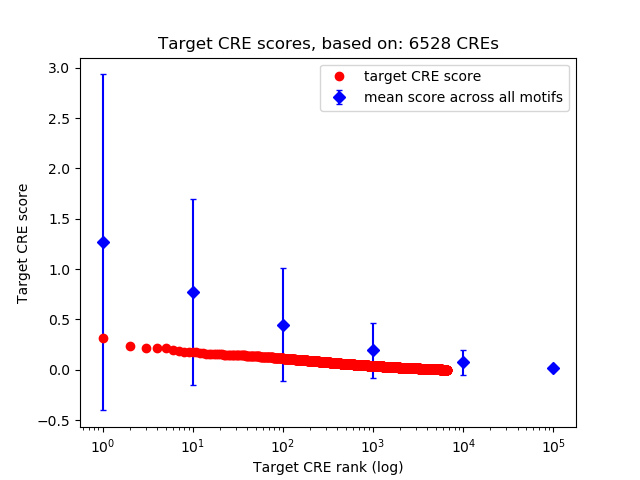
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_66159147_66159298 | HMGA2 | 58689 | 0.133634 | 0.81 | 8.2e-03 | Click! |
| chr12_66198631_66198915 | HMGA2 | 19138 | 0.213991 | 0.62 | 7.4e-02 | Click! |
| chr12_66219476_66219782 | HMGA2 | 726 | 0.709628 | 0.62 | 7.7e-02 | Click! |
| chr12_66198930_66199081 | HMGA2 | 18906 | 0.214328 | 0.62 | 7.8e-02 | Click! |
| chr12_66048530_66048681 | HMGA2 | 169306 | 0.033135 | 0.62 | 7.8e-02 | Click! |
Activity of the HMGA2 motif across conditions
Conditions sorted by the z-value of the HMGA2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
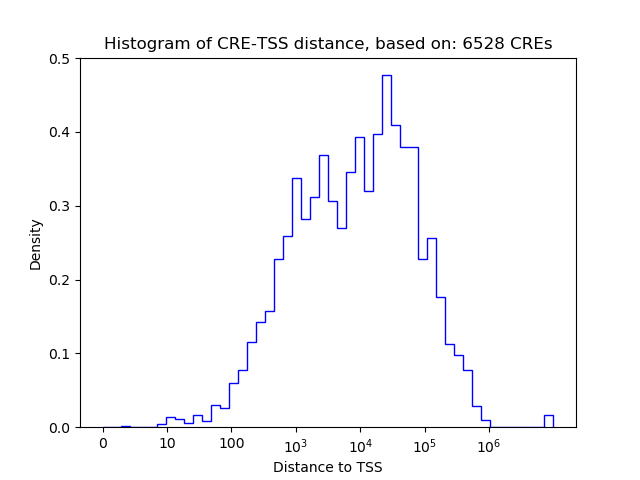
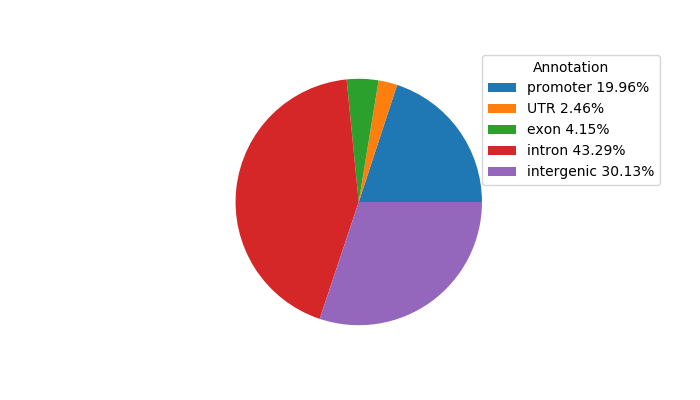
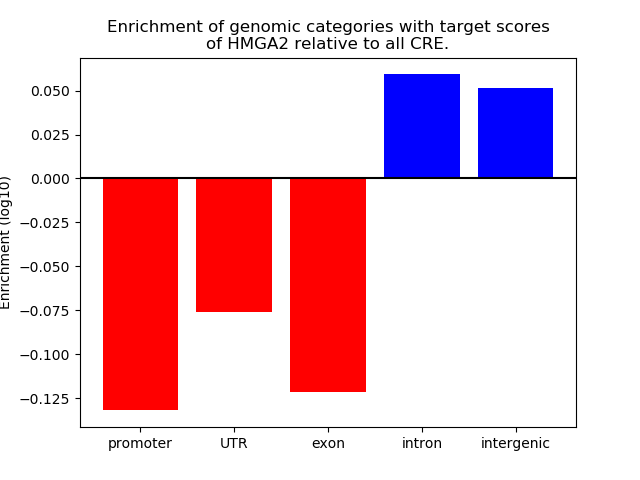
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr20_52271481_52272252 | 0.31 |
ENSG00000238468 |
. |
13431 |
0.22 |
| chr6_17986477_17986974 | 0.24 |
KIF13A |
kinesin family member 13A |
969 |
0.69 |
| chr8_53851583_53851734 | 0.21 |
NPBWR1 |
neuropeptides B/W receptor 1 |
667 |
0.79 |
| chr2_147579520_147579813 | 0.21 |
ENSG00000238860 |
. |
501873 |
0.0 |
| chr2_26100038_26101197 | 0.21 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
718 |
0.76 |
| chr4_15908125_15908316 | 0.20 |
FGFBP1 |
fibroblast growth factor binding protein 1 |
31751 |
0.17 |
| chr5_171380881_171381289 | 0.19 |
FBXW11 |
F-box and WD repeat domain containing 11 |
23675 |
0.23 |
| chr18_53255518_53256419 | 0.18 |
TCF4 |
transcription factor 4 |
108 |
0.98 |
| chr7_32929941_32930760 | 0.18 |
KBTBD2 |
kelch repeat and BTB (POZ) domain containing 2 |
415 |
0.87 |
| chr17_79371776_79372295 | 0.18 |
RP11-1055B8.7 |
BAH and coiled-coil domain-containing protein 1 |
1505 |
0.24 |
| chr4_95469395_95469810 | 0.17 |
PDLIM5 |
PDZ and LIM domain 5 |
24726 |
0.27 |
| chr3_153037931_153038307 | 0.17 |
ENSG00000265813 |
. |
120313 |
0.06 |
| chr5_139968434_139968585 | 0.17 |
ENSG00000200235 |
. |
9257 |
0.08 |
| chr10_114714851_114715858 | 0.16 |
RP11-57H14.2 |
|
3720 |
0.26 |
| chr2_188418561_188419245 | 0.16 |
AC007319.1 |
|
27 |
0.69 |
| chr12_22005080_22005298 | 0.16 |
RP11-729I10.2 |
|
25045 |
0.19 |
| chr20_14310710_14310861 | 0.16 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
7469 |
0.25 |
| chr3_27765284_27765702 | 0.15 |
EOMES |
eomesodermin |
1287 |
0.62 |
| chr18_42288728_42289100 | 0.15 |
SETBP1 |
SET binding protein 1 |
11785 |
0.32 |
| chr9_111773050_111773591 | 0.15 |
CTNNAL1 |
catenin (cadherin-associated protein), alpha-like 1 |
2459 |
0.23 |
| chr3_174819626_174819777 | 0.15 |
ENSG00000230292 |
. |
13331 |
0.3 |
| chr5_58571069_58571855 | 0.15 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
483 |
0.9 |
| chr19_6076628_6076815 | 0.15 |
CTC-232P5.3 |
|
8757 |
0.15 |
| chr4_32149379_32149669 | 0.15 |
NA |
NA |
> 106 |
NA |
| chr2_198146440_198146591 | 0.15 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
20728 |
0.16 |
| chr10_33552568_33552719 | 0.15 |
NRP1 |
neuropilin 1 |
11 |
0.98 |
| chr7_117919021_117919172 | 0.15 |
ANKRD7 |
ankyrin repeat domain 7 |
54145 |
0.17 |
| chr5_34609228_34609529 | 0.15 |
RAI14 |
retinoic acid induced 14 |
46964 |
0.16 |
| chr3_99544366_99544517 | 0.15 |
CMSS1 |
cms1 ribosomal small subunit homolog (yeast) |
7760 |
0.24 |
| chr2_40049399_40049550 | 0.15 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
35881 |
0.19 |
| chr2_238732566_238732717 | 0.15 |
RBM44 |
RNA binding motif protein 44 |
24820 |
0.17 |
| chr6_98074983_98075489 | 0.15 |
MMS22L |
MMS22-like, DNA repair protein |
344184 |
0.01 |
| chr5_153424892_153425043 | 0.15 |
MFAP3 |
microfibrillar-associated protein 3 |
6408 |
0.22 |
| chr11_37276640_37276791 | 0.14 |
ENSG00000251838 |
. |
447055 |
0.01 |
| chr4_157996824_157996975 | 0.14 |
GLRB |
glycine receptor, beta |
310 |
0.91 |
| chr10_98433378_98434045 | 0.14 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
4343 |
0.25 |
| chr6_84139529_84139855 | 0.14 |
ME1 |
malic enzyme 1, NADP(+)-dependent, cytosolic |
1072 |
0.62 |
| chr2_106886104_106886958 | 0.14 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
75736 |
0.1 |
| chr4_139162325_139164249 | 0.14 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
216 |
0.97 |
| chr16_83860165_83860316 | 0.14 |
HSBP1 |
heat shock factor binding protein 1 |
18646 |
0.15 |
| chr10_5723817_5723968 | 0.14 |
FAM208B |
family with sequence similarity 208, member B |
2909 |
0.23 |
| chr9_128511491_128511642 | 0.14 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
1088 |
0.62 |
| chr10_17241830_17242242 | 0.14 |
TRDMT1 |
tRNA aspartic acid methyltransferase 1 |
1543 |
0.37 |
| chr14_51421850_51422001 | 0.14 |
PYGL |
phosphorylase, glycogen, liver |
10471 |
0.2 |
| chr1_206685685_206685836 | 0.14 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
4881 |
0.16 |
| chr12_31470154_31471349 | 0.14 |
FAM60A |
family with sequence similarity 60, member A |
6345 |
0.16 |
| chr6_27515697_27516098 | 0.14 |
ENSG00000206671 |
. |
48294 |
0.13 |
| chr6_143140776_143141820 | 0.14 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
16886 |
0.26 |
| chr12_29716327_29716478 | 0.13 |
TMTC1 |
transmembrane and tetratricopeptide repeat containing 1 |
40755 |
0.16 |
| chr4_175135836_175136127 | 0.13 |
FBXO8 |
F-box protein 8 |
68833 |
0.12 |
| chr4_157729119_157729270 | 0.13 |
RP11-154F14.2 |
|
33317 |
0.19 |
| chr13_40081557_40081708 | 0.13 |
ENSG00000238408 |
. |
60279 |
0.13 |
| chr11_22380798_22380949 | 0.13 |
CTD-2140G10.4 |
|
2967 |
0.27 |
| chr8_13909602_13909753 | 0.13 |
RP11-3G21.1 |
|
109129 |
0.07 |
| chr9_109668731_109668882 | 0.13 |
RP11-508N12.4 |
Uncharacterized protein |
16824 |
0.21 |
| chr5_64543321_64543472 | 0.13 |
ENSG00000207439 |
. |
124200 |
0.06 |
| chr9_95590209_95590360 | 0.13 |
ENSG00000252772 |
. |
7025 |
0.17 |
| chr2_230148069_230148220 | 0.13 |
PID1 |
phosphotyrosine interaction domain containing 1 |
12143 |
0.27 |
| chr1_209528208_209528359 | 0.13 |
ENSG00000230937 |
. |
77195 |
0.12 |
| chrX_2670582_2670733 | 0.13 |
XG |
Xg blood group |
471 |
0.83 |
| chr10_75938020_75938413 | 0.13 |
ADK |
adenosine kinase |
1695 |
0.42 |
| chr3_32282148_32282299 | 0.13 |
CMTM8 |
CKLF-like MARVEL transmembrane domain containing 8 |
2052 |
0.26 |
| chr3_28025237_28025388 | 0.13 |
CMC1 |
C-x(9)-C motif containing 1 |
257774 |
0.02 |
| chr1_207489819_207490027 | 0.13 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
4930 |
0.3 |
| chr13_24782412_24782563 | 0.13 |
SPATA13 |
spermatogenesis associated 13 |
43345 |
0.13 |
| chr9_22006120_22006693 | 0.13 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
2546 |
0.22 |
| chr8_131206670_131206845 | 0.12 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
13649 |
0.23 |
| chr12_94134651_94134921 | 0.12 |
RP11-887P2.5 |
|
3187 |
0.3 |
| chr13_80913239_80913578 | 0.12 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
386 |
0.91 |
| chr21_47064838_47064989 | 0.12 |
PCBP3 |
poly(rC) binding protein 3 |
1305 |
0.55 |
| chr1_158150876_158151479 | 0.12 |
CD1D |
CD1d molecule |
1440 |
0.48 |
| chr12_109344609_109344760 | 0.12 |
ENSG00000201324 |
. |
24367 |
0.14 |
| chr1_1259044_1259204 | 0.12 |
CPSF3L |
cleavage and polyadenylation specific factor 3-like |
865 |
0.26 |
| chr10_119132010_119132432 | 0.12 |
PDZD8 |
PDZ domain containing 8 |
2757 |
0.3 |
| chr10_6779919_6780270 | 0.12 |
PRKCQ |
protein kinase C, theta |
157831 |
0.04 |
| chr17_27048575_27048801 | 0.12 |
ENSG00000238578 |
. |
912 |
0.15 |
| chr11_122177788_122178312 | 0.12 |
RP11-716H6.2 |
|
12152 |
0.26 |
| chr1_27809144_27809295 | 0.12 |
WASF2 |
WAS protein family, member 2 |
7343 |
0.18 |
| chr1_40358588_40359026 | 0.12 |
RP1-118J21.5 |
|
4610 |
0.14 |
| chr5_16701908_16702059 | 0.12 |
MYO10 |
myosin X |
36513 |
0.17 |
| chr19_1747120_1747271 | 0.12 |
ONECUT3 |
one cut homeobox 3 |
5177 |
0.15 |
| chr16_13401641_13401792 | 0.12 |
AC003009.1 |
|
24724 |
0.26 |
| chr14_85882041_85882312 | 0.12 |
RP11-497E19.2 |
Uncharacterized protein |
112767 |
0.07 |
| chr19_45147190_45148055 | 0.12 |
PVR |
poliovirus receptor |
225 |
0.9 |
| chr8_61432570_61432721 | 0.12 |
RAB2A |
RAB2A, member RAS oncogene family |
2905 |
0.3 |
| chr17_64464860_64465690 | 0.12 |
RP11-4F22.2 |
|
52303 |
0.15 |
| chr6_163838395_163838889 | 0.12 |
QKI |
QKI, KH domain containing, RNA binding |
1295 |
0.63 |
| chr16_48611180_48611331 | 0.12 |
RP11-44I10.3 |
|
17683 |
0.18 |
| chr21_34800838_34800989 | 0.12 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
3713 |
0.19 |
| chr2_58654566_58655399 | 0.12 |
FANCL |
Fanconi anemia, complementation group L |
186475 |
0.03 |
| chr1_210473057_210473208 | 0.12 |
HHAT |
hedgehog acyltransferase |
28464 |
0.19 |
| chrX_10587307_10587547 | 0.12 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
1032 |
0.65 |
| chr2_65889672_65889956 | 0.12 |
ENSG00000265899 |
. |
4576 |
0.32 |
| chr1_226846201_226846352 | 0.11 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
1095 |
0.55 |
| chrX_154300630_154300860 | 0.11 |
BRCC3 |
BRCA1/BRCA2-containing complex, subunit 3 |
941 |
0.32 |
| chr18_26737213_26737364 | 0.11 |
ENSG00000212085 |
. |
39204 |
0.19 |
| chr4_75481668_75481819 | 0.11 |
AREGB |
amphiregulin B |
1114 |
0.52 |
| chr4_149087230_149087381 | 0.11 |
RP11-76G10.1 |
|
19683 |
0.29 |
| chr5_58830530_58830681 | 0.11 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
51614 |
0.19 |
| chr4_75311912_75312063 | 0.11 |
AREG |
amphiregulin |
968 |
0.58 |
| chr13_38920865_38921016 | 0.11 |
UFM1 |
ubiquitin-fold modifier 1 |
3048 |
0.41 |
| chr16_74670457_74670608 | 0.11 |
GLG1 |
golgi glycoprotein 1 |
29520 |
0.15 |
| chr9_86955099_86955250 | 0.11 |
SLC28A3 |
solute carrier family 28 (concentrative nucleoside transporter), member 3 |
424 |
0.9 |
| chr4_83290764_83291010 | 0.11 |
HNRNPD |
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
3416 |
0.22 |
| chr1_186798918_186799215 | 0.11 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
944 |
0.73 |
| chr7_12981455_12981606 | 0.11 |
ENSG00000222974 |
. |
65221 |
0.15 |
| chrY_2620597_2620748 | 0.11 |
ENSG00000251841 |
. |
32118 |
0.19 |
| chr12_123377992_123378152 | 0.11 |
VPS37B |
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
2602 |
0.24 |
| chr9_72726451_72726602 | 0.11 |
MAMDC2-AS1 |
MAMDC2 antisense RNA 1 |
2250 |
0.38 |
| chr15_27536252_27536403 | 0.11 |
ENSG00000200326 |
. |
10744 |
0.21 |
| chr15_71035707_71036127 | 0.11 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
19852 |
0.21 |
| chr5_58592476_58593460 | 0.11 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
21023 |
0.29 |
| chr3_132316510_132316661 | 0.11 |
ACKR4 |
atypical chemokine receptor 4 |
504 |
0.83 |
| chr1_68695889_68696040 | 0.11 |
WLS |
wntless Wnt ligand secretion mediator |
1406 |
0.51 |
| chr4_54458245_54458396 | 0.11 |
LNX1 |
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
388 |
0.72 |
| chr6_27647378_27647849 | 0.11 |
ENSG00000238648 |
. |
48425 |
0.11 |
| chr7_87503109_87503295 | 0.11 |
DBF4 |
DBF4 homolog (S. cerevisiae) |
2342 |
0.29 |
| chr1_108230763_108230914 | 0.11 |
VAV3 |
vav 3 guanine nucleotide exchange factor |
288 |
0.95 |
| chr15_99416784_99417194 | 0.11 |
IGF1R |
insulin-like growth factor 1 receptor |
16581 |
0.2 |
| chr9_684288_685148 | 0.11 |
RP11-130C19.3 |
|
837 |
0.67 |
| chr9_19377459_19377736 | 0.11 |
RP11-513M16.8 |
|
2148 |
0.21 |
| chr14_91337609_91337760 | 0.10 |
TTC7B |
tetratricopeptide repeat domain 7B |
54861 |
0.15 |
| chr10_116055069_116055220 | 0.10 |
VWA2 |
von Willebrand factor A domain containing 2 |
56055 |
0.13 |
| chr16_14398873_14399280 | 0.10 |
ENSG00000207639 |
. |
1252 |
0.44 |
| chr5_76112526_76112677 | 0.10 |
F2RL1 |
coagulation factor II (thrombin) receptor-like 1 |
2157 |
0.27 |
| chr12_54090350_54090675 | 0.10 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
17901 |
0.15 |
| chr14_85999097_85999705 | 0.10 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
2829 |
0.32 |
| chr1_151432125_151432340 | 0.10 |
POGZ |
pogo transposable element with ZNF domain |
291 |
0.85 |
| chr6_86303657_86304481 | 0.10 |
SNX14 |
sorting nexin 14 |
195 |
0.95 |
| chr1_152475138_152475289 | 0.10 |
LCE5A |
late cornified envelope 5A |
8107 |
0.15 |
| chr15_59642585_59642838 | 0.10 |
ENSG00000199512 |
. |
9095 |
0.13 |
| chr6_160209552_160210274 | 0.10 |
TCP1 |
t-complex 1 |
13 |
0.95 |
| chr13_62634734_62634885 | 0.10 |
ENSG00000212007 |
. |
300300 |
0.01 |
| chr7_33080072_33080440 | 0.10 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
265 |
0.9 |
| chr6_56517425_56517576 | 0.10 |
DST |
dystonin |
9706 |
0.31 |
| chr19_14630525_14630676 | 0.10 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
1368 |
0.27 |
| chr18_52988378_52989650 | 0.10 |
TCF4 |
transcription factor 4 |
203 |
0.97 |
| chr10_21807539_21808360 | 0.10 |
SKIDA1 |
SKI/DACH domain containing 1 |
1101 |
0.4 |
| chr8_1942200_1942351 | 0.10 |
KBTBD11 |
kelch repeat and BTB (POZ) domain containing 11 |
20231 |
0.23 |
| chr5_150477046_150477197 | 0.10 |
TNIP1 |
TNFAIP3 interacting protein 1 |
3983 |
0.23 |
| chr13_80916504_80917060 | 0.10 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
1696 |
0.48 |
| chr7_25902294_25902669 | 0.10 |
ENSG00000199085 |
. |
87125 |
0.1 |
| chr5_179495571_179495863 | 0.10 |
RNF130 |
ring finger protein 130 |
3004 |
0.28 |
| chr19_46957215_46957366 | 0.10 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
17374 |
0.12 |
| chr2_72463375_72463664 | 0.10 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
88352 |
0.11 |
| chr2_135676446_135677712 | 0.10 |
CCNT2 |
cyclin T2 |
684 |
0.48 |
| chr2_187354223_187354374 | 0.10 |
ZC3H15 |
zinc finger CCCH-type containing 15 |
3318 |
0.24 |
| chr4_178495229_178495380 | 0.10 |
ENSG00000221499 |
. |
29586 |
0.22 |
| chr1_70608576_70608727 | 0.10 |
ENSG00000244389 |
. |
37472 |
0.16 |
| chr13_103424804_103424955 | 0.10 |
TEX30 |
testis expressed 30 |
1226 |
0.34 |
| chr2_101945321_101945472 | 0.10 |
ENSG00000264857 |
. |
19484 |
0.16 |
| chr10_122725495_122725646 | 0.10 |
RP11-95I16.4 |
|
85391 |
0.1 |
| chr16_65566073_65566543 | 0.10 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
410040 |
0.01 |
| chr4_157502928_157503125 | 0.10 |
RP11-171N4.2 |
Uncharacterized protein |
60433 |
0.14 |
| chr3_1830304_1830455 | 0.10 |
ENSG00000223040 |
. |
300516 |
0.01 |
| chr3_189743937_189744088 | 0.10 |
ENSG00000265045 |
. |
87711 |
0.08 |
| chr7_20372921_20373226 | 0.10 |
CTA-293F17.1 |
|
1703 |
0.35 |
| chr12_53785686_53785892 | 0.10 |
SP1 |
Sp1 transcription factor |
10703 |
0.12 |
| chr7_129425019_129425280 | 0.10 |
ENSG00000207691 |
. |
10295 |
0.15 |
| chr20_47440971_47441122 | 0.10 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
3374 |
0.34 |
| chr2_178187990_178188141 | 0.10 |
AC074286.1 |
|
173 |
0.92 |
| chr22_29280779_29281052 | 0.10 |
ZNRF3 |
zinc and ring finger 3 |
1025 |
0.55 |
| chr4_127698376_127698531 | 0.10 |
ENSG00000199862 |
. |
250921 |
0.02 |
| chr6_152085356_152085807 | 0.10 |
ESR1 |
estrogen receptor 1 |
39985 |
0.18 |
| chr3_47822404_47822881 | 0.10 |
SMARCC1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
765 |
0.62 |
| chr20_35120205_35120468 | 0.10 |
DLGAP4 |
discs, large (Drosophila) homolog-associated protein 4 |
30186 |
0.14 |
| chr9_33080885_33081164 | 0.10 |
SMU1 |
smu-1 suppressor of mec-8 and unc-52 homolog (C. elegans) |
4359 |
0.2 |
| chr17_59534634_59534785 | 0.10 |
TBX4 |
T-box 4 |
902 |
0.53 |
| chr5_114970068_114970545 | 0.10 |
TMED7 |
transmembrane emp24 protein transport domain containing 7 |
1617 |
0.33 |
| chr3_18418529_18418680 | 0.10 |
RP11-158G18.1 |
|
32523 |
0.19 |
| chr6_98566372_98566523 | 0.10 |
ENSG00000238367 |
. |
94040 |
0.1 |
| chr11_12824646_12824913 | 0.10 |
RP11-47J17.3 |
|
20435 |
0.2 |
| chr12_84188120_84188271 | 0.10 |
ENSG00000221148 |
. |
389032 |
0.01 |
| chr15_62599027_62599178 | 0.10 |
RP11-299H22.5 |
|
36223 |
0.17 |
| chr1_164652386_164652537 | 0.10 |
RP11-477H21.2 |
|
3139 |
0.29 |
| chr5_149857239_149857390 | 0.10 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
8067 |
0.18 |
| chr16_82172666_82172817 | 0.10 |
RP11-510J16.5 |
|
495 |
0.79 |
| chr6_28175291_28175856 | 0.10 |
ZSCAN9 |
zinc finger and SCAN domain containing 9 |
17091 |
0.12 |
| chr15_61963769_61963920 | 0.09 |
ENSG00000266349 |
. |
72296 |
0.13 |
| chr3_107806842_107807168 | 0.09 |
CD47 |
CD47 molecule |
2856 |
0.4 |
| chr3_142607235_142608274 | 0.09 |
PCOLCE2 |
procollagen C-endopeptidase enhancer 2 |
13 |
0.98 |
| chr10_33251129_33251359 | 0.09 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
967 |
0.68 |
| chr11_93123532_93123683 | 0.09 |
CCDC67 |
coiled-coil domain containing 67 |
6973 |
0.23 |
| chr1_10511739_10511890 | 0.09 |
CORT |
cortistatin |
1843 |
0.2 |
| chr20_52516714_52516865 | 0.09 |
AC005220.3 |
|
39910 |
0.19 |
| chr5_142585460_142585745 | 0.09 |
ARHGAP26 |
Rho GTPase activating protein 26 |
1161 |
0.58 |
| chr10_103813684_103813835 | 0.09 |
C10orf76 |
chromosome 10 open reading frame 76 |
2191 |
0.26 |
| chr2_212272854_212273005 | 0.09 |
ENSG00000199585 |
. |
127449 |
0.06 |
| chr1_152010543_152010714 | 0.09 |
S100A11 |
S100 calcium binding protein A11 |
1117 |
0.44 |
| chr22_36233928_36234199 | 0.09 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
2202 |
0.43 |
| chr1_10124465_10124616 | 0.09 |
UBE4B |
ubiquitination factor E4B |
30886 |
0.14 |
| chr4_120987081_120987232 | 0.09 |
RP11-679C8.2 |
|
957 |
0.45 |
| chr17_75128680_75128831 | 0.09 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
4800 |
0.22 |
| chr5_38854613_38854764 | 0.09 |
OSMR |
oncostatin M receptor |
8587 |
0.28 |
| chrX_31477449_31477600 | 0.09 |
ENSG00000252903 |
. |
111215 |
0.07 |
| chr14_89507425_89507846 | 0.09 |
FOXN3 |
forkhead box N3 |
139453 |
0.05 |
| chr1_158123360_158123511 | 0.09 |
CD1D |
CD1d molecule |
26302 |
0.19 |
| chr9_75566785_75567046 | 0.09 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
1056 |
0.7 |
| chr21_38081043_38081194 | 0.09 |
SIM2 |
single-minded family bHLH transcription factor 2 |
363 |
0.88 |
| chr7_18549761_18549974 | 0.09 |
HDAC9 |
histone deacetylase 9 |
931 |
0.71 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.0 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.0 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.0 | GO:0043302 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.0 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.0 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:1903170 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:0090267 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |