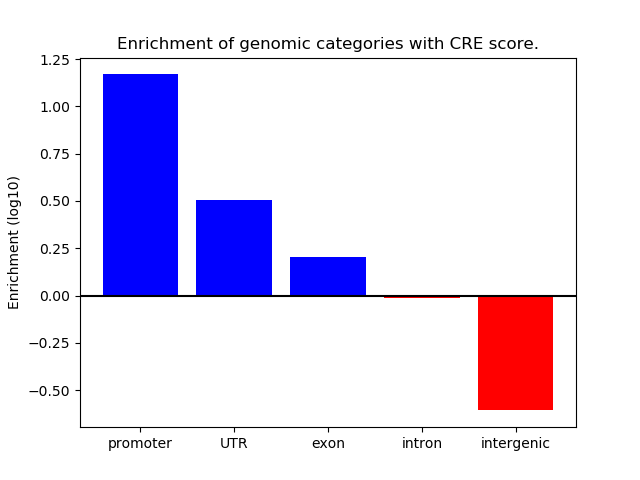
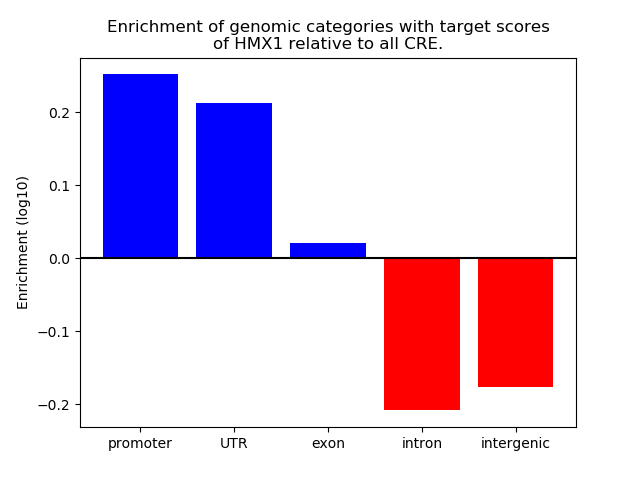
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
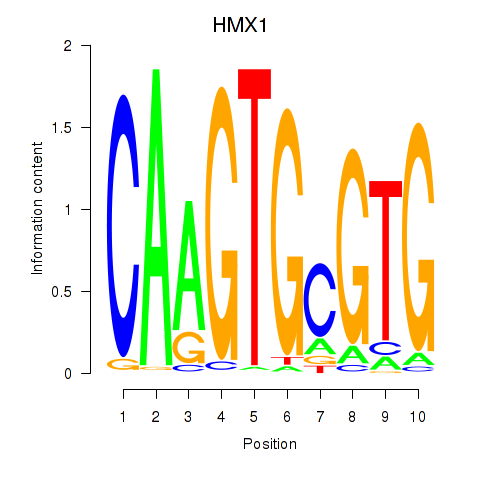
Results for HMX1
Z-value: 0.41
Transcription factors associated with HMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX1
|
ENSG00000215612.5 | H6 family homeobox 1 |
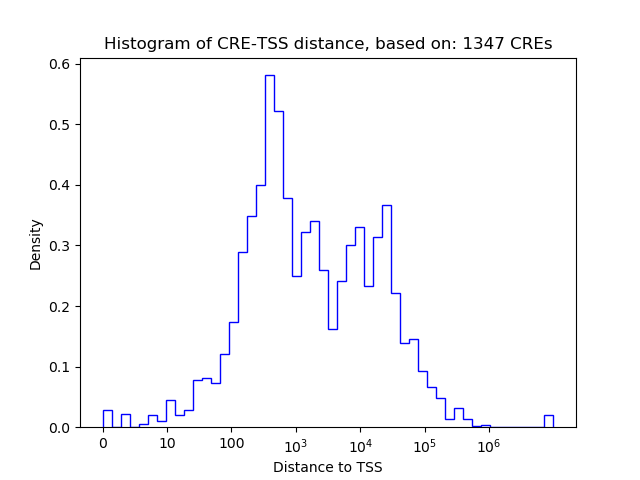
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_8940977_8941128 | HMX1 | 67509 | 0.098319 | -0.40 | 2.8e-01 | Click! |
Activity of the HMX1 motif across conditions
Conditions sorted by the z-value of the HMX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
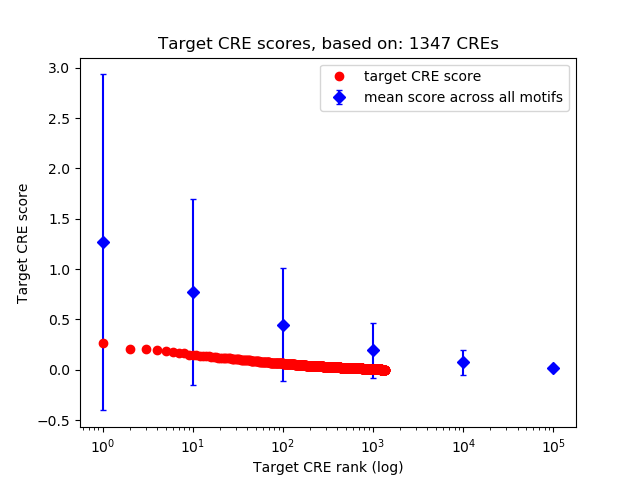
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_175497594_175497986 | 0.26 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
1517 |
0.45 |
| chr17_62097338_62097710 | 0.21 |
ICAM2 |
intercellular adhesion molecule 2 |
387 |
0.83 |
| chr17_76823234_76823385 | 0.20 |
USP36 |
ubiquitin specific peptidase 36 |
1737 |
0.32 |
| chr19_54815773_54815924 | 0.20 |
LILRA3 |
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 |
5896 |
0.1 |
| chr2_144855004_144855155 | 0.18 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
138225 |
0.05 |
| chr22_20818482_20818633 | 0.17 |
ENSG00000255156 |
. |
11041 |
0.09 |
| chr11_128392558_128393039 | 0.17 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
593 |
0.77 |
| chr14_101586704_101586913 | 0.17 |
ENSG00000206761 |
. |
25224 |
0.04 |
| chr22_37881738_37882044 | 0.15 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
534 |
0.71 |
| chr12_133346878_133347204 | 0.15 |
ANKLE2 |
ankyrin repeat and LEM domain containing 2 |
8567 |
0.15 |
| chr5_90678270_90678948 | 0.14 |
ARRDC3 |
arrestin domain containing 3 |
567 |
0.83 |
| chr6_53200265_53200465 | 0.14 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
13222 |
0.2 |
| chr11_3849313_3849464 | 0.14 |
RHOG |
ras homolog family member G |
2493 |
0.15 |
| chr9_132648514_132648803 | 0.14 |
FNBP1 |
formin binding protein 1 |
32931 |
0.12 |
| chr1_192130381_192130596 | 0.13 |
RGS18 |
regulator of G-protein signaling 18 |
2901 |
0.41 |
| chr1_28503293_28503444 | 0.13 |
PTAFR |
platelet-activating factor receptor |
331 |
0.81 |
| chr17_56412846_56413076 | 0.13 |
ENSG00000264399 |
. |
422 |
0.69 |
| chr16_57334462_57334613 | 0.13 |
PLLP |
plasmolipin |
15938 |
0.14 |
| chr7_106505707_106506635 | 0.12 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
247 |
0.95 |
| chr11_47397838_47397989 | 0.12 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
2029 |
0.17 |
| chr1_160614614_160614765 | 0.12 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
2122 |
0.27 |
| chr9_140131231_140131459 | 0.12 |
TUBB4B |
tubulin, beta 4B class IVb |
4320 |
0.07 |
| chr12_4382982_4383628 | 0.12 |
CCND2 |
cyclin D2 |
367 |
0.72 |
| chr2_85906566_85906717 | 0.12 |
SFTPB |
surfactant protein B |
10777 |
0.13 |
| chr10_54514994_54515145 | 0.11 |
RP11-556E13.1 |
|
100 |
0.98 |
| chr15_86087225_86087376 | 0.11 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
29 |
0.98 |
| chr1_244979206_244979357 | 0.11 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
19343 |
0.18 |
| chr1_149754304_149754780 | 0.11 |
FCGR1A |
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
296 |
0.78 |
| chr1_149369357_149369823 | 0.11 |
FCGR1C |
Fc fragment of IgG, high affinity Ic, receptor (CD64), pseudogene |
233 |
0.9 |
| chr10_51566009_51566382 | 0.11 |
NCOA4 |
nuclear receptor coactivator 4 |
965 |
0.54 |
| chr16_29638502_29638653 | 0.10 |
ENSG00000266758 |
. |
27991 |
0.11 |
| chr8_142219382_142219533 | 0.10 |
DENND3 |
DENN/MADD domain containing 3 |
17027 |
0.15 |
| chr11_65307798_65308115 | 0.10 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
33 |
0.95 |
| chr14_105530971_105531214 | 0.10 |
GPR132 |
G protein-coupled receptor 132 |
675 |
0.7 |
| chr19_54711508_54711701 | 0.10 |
RPS9 |
ribosomal protein S9 |
6576 |
0.08 |
| chr17_81146893_81147147 | 0.10 |
METRNL |
meteorin, glial cell differentiation regulator-like |
95026 |
0.08 |
| chr17_43301087_43301318 | 0.10 |
CTD-2020K17.1 |
|
1613 |
0.2 |
| chr7_105332169_105332578 | 0.10 |
ATXN7L1 |
ataxin 7-like 1 |
289 |
0.94 |
| chrX_40869304_40869484 | 0.10 |
USP9X |
ubiquitin specific peptidase 9, X-linked |
75494 |
0.12 |
| chr1_120935370_120935573 | 0.10 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
423 |
0.82 |
| chr2_38607378_38607679 | 0.09 |
ATL2 |
atlastin GTPase 2 |
3101 |
0.37 |
| chr17_16117665_16118816 | 0.09 |
NCOR1 |
nuclear receptor corepressor 1 |
603 |
0.66 |
| chr22_17082773_17083116 | 0.09 |
ENSG00000221084 |
. |
3250 |
0.22 |
| chr19_47237277_47237428 | 0.09 |
ENSG00000222614 |
. |
2217 |
0.15 |
| chr5_126144250_126144473 | 0.09 |
LMNB1 |
lamin B1 |
31480 |
0.19 |
| chrY_289482_289633 | 0.09 |
NA |
NA |
> 106 |
NA |
| chr17_37558067_37558428 | 0.09 |
CTB-131K11.1 |
|
201 |
0.65 |
| chr18_9914707_9915136 | 0.09 |
VAPA |
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
862 |
0.58 |
| chr6_10528716_10529334 | 0.09 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
436 |
0.83 |
| chr17_75878240_75878391 | 0.09 |
FLJ45079 |
|
344 |
0.91 |
| chr10_103875323_103875595 | 0.09 |
LDB1 |
LIM domain binding 1 |
756 |
0.56 |
| chr11_65695547_65695843 | 0.09 |
DRAP1 |
DR1-associated protein 1 (negative cofactor 2 alpha) |
8500 |
0.09 |
| chr6_22063278_22063579 | 0.08 |
ENSG00000222515 |
. |
22345 |
0.28 |
| chr4_118925834_118925985 | 0.08 |
NDST3 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
29591 |
0.26 |
| chr9_129295639_129295790 | 0.08 |
ENSG00000221768 |
. |
2444 |
0.31 |
| chr11_128564573_128565732 | 0.08 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
766 |
0.51 |
| chr7_102066305_102066456 | 0.08 |
ORAI2 |
ORAI calcium release-activated calcium modulator 2 |
7173 |
0.09 |
| chr11_62361155_62361306 | 0.08 |
TUT1 |
terminal uridylyl transferase 1, U6 snRNA-specific |
1581 |
0.16 |
| chr17_66379730_66379881 | 0.08 |
ENSG00000207561 |
. |
40884 |
0.14 |
| chr10_101945955_101946507 | 0.08 |
ERLIN1 |
ER lipid raft associated 1 |
441 |
0.76 |
| chr21_36420108_36421159 | 0.08 |
RUNX1 |
runt-related transcription factor 1 |
829 |
0.77 |
| chr3_122477747_122477898 | 0.08 |
ENSG00000238480 |
. |
8603 |
0.19 |
| chr19_41329914_41330065 | 0.08 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
16841 |
0.11 |
| chr3_53204570_53204956 | 0.08 |
PRKCD |
protein kinase C, delta |
5618 |
0.19 |
| chr2_242679805_242679956 | 0.08 |
D2HGDH |
D-2-hydroxyglutarate dehydrogenase |
1968 |
0.18 |
| chr16_11038476_11038796 | 0.08 |
CLEC16A |
C-type lectin domain family 16, member A |
198 |
0.9 |
| chr20_30639783_30640305 | 0.08 |
HCK |
hemopoietic cell kinase |
1 |
0.96 |
| chr10_134369040_134369355 | 0.08 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
17554 |
0.21 |
| chr17_73772737_73772888 | 0.07 |
H3F3B |
H3 histone, family 3B (H3.3B) |
2555 |
0.14 |
| chrX_48775697_48776218 | 0.07 |
PIM2 |
pim-2 oncogene |
344 |
0.77 |
| chr7_2491547_2491962 | 0.07 |
CHST12 |
carbohydrate (chondroitin 4) sulfotransferase 12 |
48020 |
0.11 |
| chr9_95726568_95726886 | 0.07 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
484 |
0.84 |
| chr5_77147504_77148481 | 0.07 |
TBCA |
tubulin folding cofactor A |
16612 |
0.28 |
| chr12_2453066_2453260 | 0.07 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
74221 |
0.11 |
| chr19_8639062_8639292 | 0.07 |
MYO1F |
myosin IF |
3145 |
0.16 |
| chr2_99082083_99082234 | 0.07 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
20745 |
0.22 |
| chr1_208712376_208712638 | 0.07 |
PLXNA2 |
plexin A2 |
294842 |
0.01 |
| chr17_76255043_76255413 | 0.07 |
TMEM235 |
transmembrane protein 235 |
27106 |
0.12 |
| chr16_84421_84572 | 0.07 |
WASIR2 |
WASH and IL9R antisense RNA 2 |
11586 |
0.1 |
| chr19_45621503_45621654 | 0.07 |
AC005757.6 |
|
18657 |
0.08 |
| chr8_134086953_134087155 | 0.07 |
SLA |
Src-like-adaptor |
14451 |
0.23 |
| chr19_51728373_51728524 | 0.07 |
CD33 |
CD33 molecule |
94 |
0.94 |
| chr17_19883077_19883228 | 0.07 |
AKAP10 |
A kinase (PRKA) anchor protein 10 |
2002 |
0.3 |
| chr14_58666937_58667480 | 0.07 |
ACTR10 |
actin-related protein 10 homolog (S. cerevisiae) |
352 |
0.84 |
| chr17_4389201_4389708 | 0.07 |
RP13-580F15.2 |
|
194 |
0.92 |
| chr4_80993397_80993717 | 0.07 |
ANTXR2 |
anthrax toxin receptor 2 |
160 |
0.97 |
| chr19_3178230_3178381 | 0.06 |
S1PR4 |
sphingosine-1-phosphate receptor 4 |
431 |
0.74 |
| chr6_88973481_88973632 | 0.06 |
CNR1 |
cannabinoid receptor 1 (brain) |
97478 |
0.09 |
| chr21_37572727_37572878 | 0.06 |
ENSG00000265882 |
. |
13334 |
0.14 |
| chr22_17700031_17700512 | 0.06 |
CECR1 |
cat eye syndrome chromosome region, candidate 1 |
5 |
0.98 |
| chr11_9285524_9286554 | 0.06 |
DENND5A |
DENN/MADD domain containing 5A |
595 |
0.74 |
| chr6_37017159_37017310 | 0.06 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
4627 |
0.23 |
| chr15_73928751_73928902 | 0.06 |
NPTN |
neuroplastin |
2351 |
0.36 |
| chr1_236105367_236105518 | 0.06 |
ENSG00000206803 |
. |
26727 |
0.17 |
| chr16_29984700_29984855 | 0.06 |
TAOK2 |
TAO kinase 2 |
185 |
0.87 |
| chr20_62434745_62434896 | 0.06 |
RP4-583P15.11 |
|
4609 |
0.11 |
| chr13_49065462_49065706 | 0.06 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
41629 |
0.17 |
| chr3_151987094_151987576 | 0.06 |
MBNL1-AS1 |
MBNL1 antisense RNA 1 |
0 |
0.58 |
| chr1_160990016_160990876 | 0.06 |
F11R |
F11 receptor |
440 |
0.71 |
| chr16_30583519_30583791 | 0.06 |
ZNF688 |
zinc finger protein 688 |
73 |
0.49 |
| chr1_54821806_54822159 | 0.06 |
SSBP3 |
single stranded DNA binding protein 3 |
49195 |
0.13 |
| chrX_37640522_37640673 | 0.06 |
CYBB |
cytochrome b-245, beta polypeptide |
1280 |
0.54 |
| chr8_22563265_22563416 | 0.06 |
EGR3 |
early growth response 3 |
12525 |
0.13 |
| chr10_45464087_45464238 | 0.06 |
RASSF4 |
Ras association (RalGDS/AF-6) domain family member 4 |
1510 |
0.28 |
| chr2_202097823_202098101 | 0.06 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
204 |
0.94 |
| chr9_71319950_71320419 | 0.06 |
PIP5K1B |
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
391 |
0.9 |
| chr1_27147150_27147301 | 0.06 |
ENSG00000241188 |
. |
5912 |
0.12 |
| chr12_129280330_129280481 | 0.06 |
SLC15A4 |
solute carrier family 15 (oligopeptide transporter), member 4 |
5841 |
0.28 |
| chr12_40618871_40619488 | 0.06 |
LRRK2 |
leucine-rich repeat kinase 2 |
303 |
0.89 |
| chr11_6640777_6640928 | 0.06 |
TPP1 |
tripeptidyl peptidase I |
160 |
0.82 |
| chr20_23028362_23028696 | 0.06 |
THBD |
thrombomodulin |
1849 |
0.28 |
| chr14_93579935_93580162 | 0.06 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
1569 |
0.39 |
| chr12_125052371_125052867 | 0.06 |
NCOR2 |
nuclear receptor corepressor 2 |
609 |
0.84 |
| chr13_52212503_52212693 | 0.06 |
ENSG00000242893 |
. |
44247 |
0.11 |
| chr17_39655103_39655254 | 0.06 |
AC019349.5 |
|
2057 |
0.14 |
| chr14_91830192_91830394 | 0.06 |
ENSG00000265856 |
. |
30236 |
0.18 |
| chrX_44173865_44174016 | 0.06 |
EFHC2 |
EF-hand domain (C-terminal) containing 2 |
28978 |
0.24 |
| chr20_4794055_4794206 | 0.06 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
1639 |
0.4 |
| chr6_110011634_110011785 | 0.06 |
AK9 |
adenylate kinase 9 |
0 |
0.95 |
| chr12_125478431_125478707 | 0.06 |
BRI3BP |
BRI3 binding protein |
323 |
0.89 |
| chr7_5862773_5862988 | 0.06 |
ZNF815P |
zinc finger protein 815, pseudogene |
42 |
0.97 |
| chr3_112686562_112686713 | 0.06 |
CD200R1 |
CD200 receptor 1 |
7122 |
0.17 |
| chr20_62714432_62714977 | 0.06 |
C20orf201 |
chromosome 20 open reading frame 201 |
1008 |
0.3 |
| chr15_51913253_51913493 | 0.06 |
DMXL2 |
Dmx-like 2 |
1459 |
0.4 |
| chr1_145437718_145438228 | 0.06 |
TXNIP |
thioredoxin interacting protein |
496 |
0.69 |
| chr3_4536182_4536507 | 0.06 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
1110 |
0.44 |
| chr1_153917802_153918374 | 0.06 |
DENND4B |
DENN/MADD domain containing 4B |
343 |
0.75 |
| chr12_8234042_8234677 | 0.06 |
NECAP1 |
NECAP endocytosis associated 1 |
448 |
0.78 |
| chr9_4942396_4942547 | 0.06 |
ENSG00000238362 |
. |
25626 |
0.17 |
| chr7_135656031_135656246 | 0.05 |
MTPN |
myotrophin |
5926 |
0.18 |
| chr19_10748205_10748356 | 0.05 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
40 |
0.96 |
| chr6_32156796_32157600 | 0.05 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
765 |
0.38 |
| chr2_171571264_171571992 | 0.05 |
SP5 |
Sp5 transcription factor |
233 |
0.76 |
| chr15_67356717_67357002 | 0.05 |
SMAD3 |
SMAD family member 3 |
758 |
0.77 |
| chr19_49243178_49243445 | 0.05 |
RASIP1 |
Ras interacting protein 1 |
667 |
0.48 |
| chr4_38784523_38784674 | 0.05 |
TLR10 |
toll-like receptor 10 |
9 |
0.97 |
| chr16_88878512_88878743 | 0.05 |
APRT |
adenine phosphoribosyltransferase |
275 |
0.82 |
| chr17_76381899_76382050 | 0.05 |
PGS1 |
phosphatidylglycerophosphate synthase 1 |
7215 |
0.14 |
| chr7_29628654_29628805 | 0.05 |
PRR15 |
proline rich 15 |
23663 |
0.17 |
| chr17_72527307_72527458 | 0.05 |
CD300LB |
CD300 molecule-like family member b |
223 |
0.91 |
| chr17_38486750_38487092 | 0.05 |
RARA |
retinoic acid receptor, alpha |
10719 |
0.11 |
| chr12_102090642_102091223 | 0.05 |
CHPT1 |
choline phosphotransferase 1 |
473 |
0.75 |
| chr3_126191945_126192170 | 0.05 |
ZXDC |
ZXD family zinc finger C |
2651 |
0.25 |
| chr5_37370451_37371178 | 0.05 |
NUP155 |
nucleoporin 155kDa |
73 |
0.98 |
| chr6_7145795_7146333 | 0.05 |
RREB1 |
ras responsive element binding protein 1 |
6586 |
0.22 |
| chr19_2290091_2290388 | 0.05 |
LINGO3 |
leucine rich repeat and Ig domain containing 3 |
1784 |
0.18 |
| chr6_5026001_5026185 | 0.05 |
ENSG00000252419 |
. |
20242 |
0.19 |
| chr1_202457690_202457841 | 0.05 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
25894 |
0.17 |
| chr11_59554604_59554755 | 0.05 |
STX3 |
syntaxin 3 |
139 |
0.95 |
| chr17_43334091_43334242 | 0.05 |
MAP3K14-AS1 |
MAP3K14 antisense RNA 1 |
1775 |
0.2 |
| chrX_153284800_153285224 | 0.05 |
IRAK1 |
interleukin-1 receptor-associated kinase 1 |
239 |
0.66 |
| chr14_70104707_70105002 | 0.05 |
KIAA0247 |
KIAA0247 |
26541 |
0.2 |
| chr1_206729975_206731239 | 0.05 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
114 |
0.96 |
| chr12_65004453_65004740 | 0.05 |
RASSF3 |
Ras association (RalGDS/AF-6) domain family member 3 |
303 |
0.81 |
| chr3_71631883_71632034 | 0.05 |
FOXP1 |
forkhead box P1 |
946 |
0.6 |
| chr15_75077997_75078148 | 0.05 |
CSK |
c-src tyrosine kinase |
2813 |
0.16 |
| chr16_28996496_28996848 | 0.05 |
LAT |
linker for activation of T cells |
11 |
0.94 |
| chr14_98108128_98108342 | 0.05 |
ENSG00000240730 |
. |
111725 |
0.07 |
| chr19_3505802_3505997 | 0.05 |
FZR1 |
fizzy/cell division cycle 20 related 1 (Drosophila) |
372 |
0.73 |
| chr19_2096024_2096259 | 0.05 |
MOB3A |
MOB kinase activator 3A |
128 |
0.64 |
| chr1_167522333_167523011 | 0.05 |
CREG1 |
cellular repressor of E1A-stimulated genes 1 |
332 |
0.9 |
| chr1_92942181_92942332 | 0.05 |
GFI1 |
growth factor independent 1 transcription repressor |
7255 |
0.27 |
| chr11_111741428_111741733 | 0.05 |
ALG9 |
ALG9, alpha-1,2-mannosyltransferase |
8 |
0.96 |
| chr3_53736497_53736648 | 0.05 |
CACNA1D |
calcium channel, voltage-dependent, L type, alpha 1D subunit |
36144 |
0.17 |
| chr2_67610232_67610383 | 0.05 |
ETAA1 |
Ewing tumor-associated antigen 1 |
14144 |
0.32 |
| chr15_91372562_91372713 | 0.05 |
CTD-3094K11.1 |
|
10317 |
0.13 |
| chr12_52294368_52294519 | 0.05 |
ACVRL1 |
activin A receptor type II-like 1 |
6249 |
0.18 |
| chr21_29782788_29782939 | 0.04 |
ENSG00000251894 |
. |
332667 |
0.01 |
| chr13_25085743_25086881 | 0.04 |
PARP4 |
poly (ADP-ribose) polymerase family, member 4 |
636 |
0.74 |
| chr11_65292001_65292182 | 0.04 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
457 |
0.69 |
| chr6_13434336_13434487 | 0.04 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
26042 |
0.17 |
| chr6_2901280_2901469 | 0.04 |
SERPINB9 |
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
2140 |
0.3 |
| chr12_6447373_6447724 | 0.04 |
TNFRSF1A |
tumor necrosis factor receptor superfamily, member 1A |
3466 |
0.15 |
| chr6_149771705_149771910 | 0.04 |
ZC3H12D |
zinc finger CCCH-type containing 12D |
6198 |
0.19 |
| chr10_90593250_90593401 | 0.04 |
ANKRD22 |
ankyrin repeat domain 22 |
18250 |
0.16 |
| chr1_235097453_235097604 | 0.04 |
ENSG00000239690 |
. |
57595 |
0.14 |
| chr15_81591495_81592151 | 0.04 |
IL16 |
interleukin 16 |
66 |
0.98 |
| chr15_65117601_65117801 | 0.04 |
PIF1 |
PIF1 5'-to-3' DNA helicase |
137 |
0.94 |
| chr14_56584316_56584807 | 0.04 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
29 |
0.98 |
| chr21_46347709_46348073 | 0.04 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
742 |
0.47 |
| chr8_142384090_142384264 | 0.04 |
GPR20 |
G protein-coupled receptor 20 |
6810 |
0.12 |
| chr10_31073473_31074431 | 0.04 |
RP11-330O11.3 |
|
48364 |
0.15 |
| chrX_71341466_71341617 | 0.04 |
RGAG4 |
retrotransposon gag domain containing 4 |
10137 |
0.16 |
| chr11_5646501_5646907 | 0.04 |
TRIM34 |
tripartite motif containing 34 |
491 |
0.45 |
| chr3_4535116_4535869 | 0.04 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
258 |
0.78 |
| chr11_61659243_61659612 | 0.04 |
FADS3 |
fatty acid desaturase 3 |
96 |
0.95 |
| chr19_50400763_50401028 | 0.04 |
IL4I1 |
interleukin 4 induced 1 |
683 |
0.39 |
| chr11_128595318_128595476 | 0.04 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
29479 |
0.16 |
| chr3_138594988_138595139 | 0.04 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
41283 |
0.15 |
| chr22_37310592_37310811 | 0.04 |
CSF2RB |
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
1026 |
0.44 |
| chr17_72588244_72588395 | 0.04 |
CD300LD |
CD300 molecule-like family member d |
103 |
0.94 |
| chr1_65885426_65885920 | 0.04 |
LEPR |
leptin receptor |
575 |
0.45 |
| chr22_37639776_37640249 | 0.04 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
276 |
0.88 |
| chr15_41695201_41695405 | 0.04 |
NDUFAF1 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
586 |
0.69 |
| chr2_10091778_10091990 | 0.04 |
GRHL1 |
grainyhead-like 1 (Drosophila) |
57 |
0.98 |
| chr6_53109316_53109571 | 0.04 |
ENSG00000206908 |
. |
25384 |
0.15 |
| chr17_47840030_47840441 | 0.04 |
FAM117A |
family with sequence similarity 117, member A |
1258 |
0.41 |
| chr14_76620742_76620893 | 0.04 |
GPATCH2L |
G patch domain containing 2-like |
828 |
0.72 |
| chr1_206731858_206732141 | 0.04 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
1506 |
0.37 |
| chr3_13056943_13057225 | 0.04 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
28548 |
0.23 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0006168 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |