Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
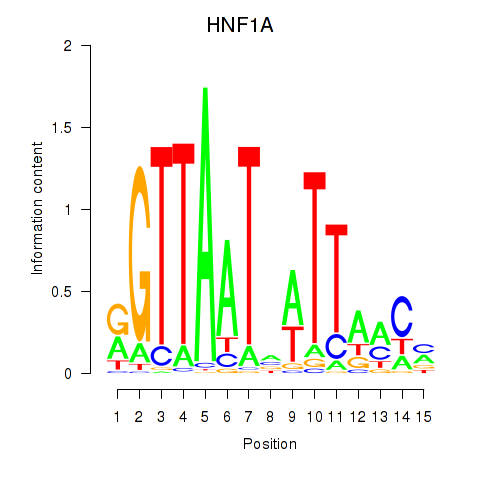
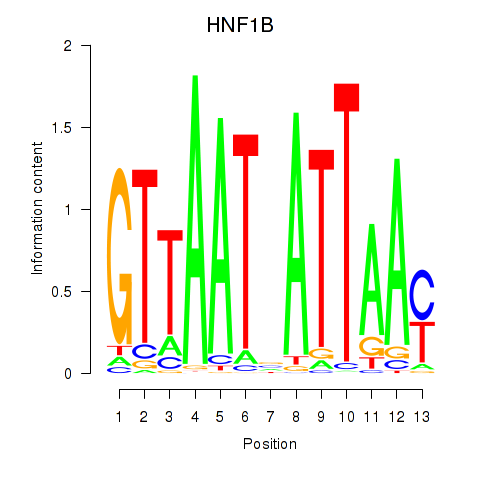
Results for HNF1A_HNF1B
Z-value: 0.64
Transcription factors associated with HNF1A_HNF1B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF1A
|
ENSG00000135100.13 | HNF1 homeobox A |
|
HNF1B
|
ENSG00000108753.8 | HNF1 homeobox B |
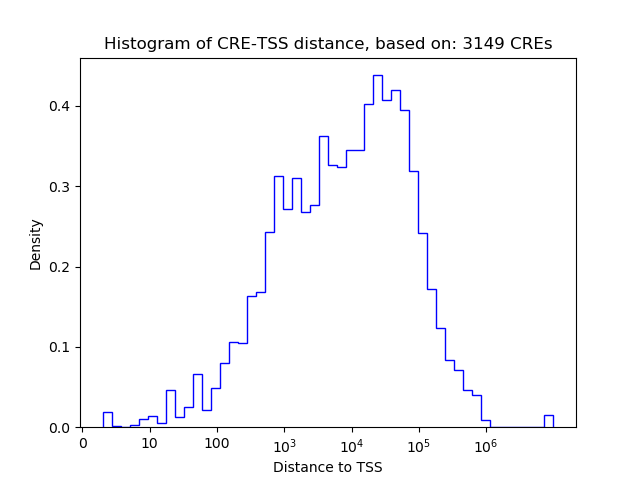
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_36102015_36102652 | HNF1B | 2676 | 0.245172 | 0.75 | 1.9e-02 | Click! |
| chr17_36102983_36103168 | HNF1B | 1934 | 0.287380 | 0.67 | 4.9e-02 | Click! |
| chr17_36102697_36102848 | HNF1B | 2237 | 0.266462 | 0.60 | 8.9e-02 | Click! |
| chr17_36104012_36104163 | HNF1B | 922 | 0.435270 | 0.54 | 1.3e-01 | Click! |
| chr17_36104676_36104827 | HNF1B | 258 | 0.681643 | 0.44 | 2.3e-01 | Click! |
Activity of the HNF1A_HNF1B motif across conditions
Conditions sorted by the z-value of the HNF1A_HNF1B motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
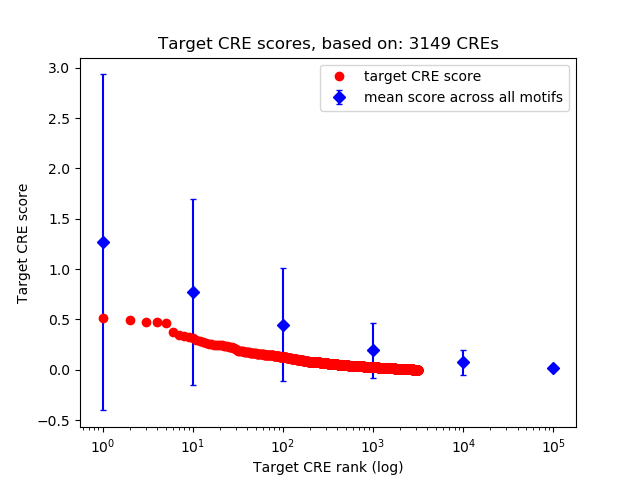
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_85768780_85768931 | 0.51 |
ALX1 |
ALX homeobox 1 |
94970 |
0.09 |
| chr18_20739662_20739813 | 0.49 |
CABLES1 |
Cdk5 and Abl enzyme substrate 1 |
3948 |
0.25 |
| chr1_200865204_200865355 | 0.48 |
C1orf106 |
chromosome 1 open reading frame 106 |
1330 |
0.42 |
| chr5_101774760_101774911 | 0.47 |
SLCO6A1 |
solute carrier organic anion transporter family, member 6A1 |
59782 |
0.13 |
| chr2_133174054_133174205 | 0.47 |
GPR39 |
G protein-coupled receptor 39 |
18 |
0.98 |
| chr10_11602391_11602542 | 0.38 |
USP6NL |
USP6 N-terminal like |
13270 |
0.25 |
| chr6_12364096_12364247 | 0.35 |
ENSG00000223321 |
. |
42842 |
0.19 |
| chr11_16946345_16946496 | 0.34 |
PLEKHA7 |
pleckstrin homology domain containing, family A member 7 |
43515 |
0.12 |
| chr1_211688650_211688892 | 0.33 |
RD3 |
retinal degeneration 3 |
22512 |
0.2 |
| chr18_21458592_21458743 | 0.31 |
LAMA3 |
laminin, alpha 3 |
5658 |
0.23 |
| chr3_111452039_111452190 | 0.30 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
770 |
0.71 |
| chr3_183947999_183948171 | 0.29 |
VWA5B2 |
von Willebrand factor A domain containing 5B2 |
132 |
0.92 |
| chr10_46010179_46010570 | 0.27 |
MARCH8 |
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
20445 |
0.22 |
| chr1_97344163_97344314 | 0.26 |
ENSG00000212039 |
. |
24017 |
0.23 |
| chr17_37895484_37895662 | 0.26 |
GRB7 |
growth factor receptor-bound protein 7 |
549 |
0.64 |
| chr12_107991801_107991952 | 0.25 |
BTBD11 |
BTB (POZ) domain containing 11 |
17449 |
0.18 |
| chr2_234601388_234601539 | 0.25 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
49 |
0.95 |
| chr3_47822404_47822881 | 0.24 |
SMARCC1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
765 |
0.62 |
| chr3_33138761_33138999 | 0.24 |
GLB1 |
galactosidase, beta 1 |
158 |
0.84 |
| chr10_120966033_120966362 | 0.24 |
GRK5 |
G protein-coupled receptor kinase 5 |
904 |
0.45 |
| chr22_25508803_25509083 | 0.24 |
CTA-221G9.11 |
|
284 |
0.93 |
| chr1_207204103_207204254 | 0.24 |
C1orf116 |
chromosome 1 open reading frame 116 |
1914 |
0.24 |
| chr14_61188910_61189596 | 0.24 |
SIX4 |
SIX homeobox 4 |
1599 |
0.41 |
| chr8_118959706_118959909 | 0.23 |
EXT1 |
exostosin glycosyltransferase 1 |
162846 |
0.04 |
| chr13_77466816_77466967 | 0.23 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
6351 |
0.21 |
| chr9_20320242_20320393 | 0.22 |
ENSG00000221744 |
. |
25315 |
0.21 |
| chr19_12722754_12723067 | 0.22 |
ZNF791 |
zinc finger protein 791 |
1142 |
0.25 |
| chr4_75818567_75818718 | 0.22 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
39663 |
0.18 |
| chr11_2405249_2405453 | 0.22 |
CD81 |
CD81 molecule |
529 |
0.65 |
| chr10_29924729_29924934 | 0.20 |
SVIL |
supervillin |
930 |
0.62 |
| chr1_246324865_246325016 | 0.20 |
ENSG00000202184 |
. |
27480 |
0.18 |
| chr9_74909575_74909726 | 0.19 |
ENSG00000252428 |
. |
5104 |
0.28 |
| chr14_58748738_58749043 | 0.19 |
RP11-349A22.5 |
|
4851 |
0.12 |
| chr1_6420005_6420156 | 0.19 |
ACOT7 |
acyl-CoA thioesterase 7 |
676 |
0.64 |
| chr18_9648789_9648940 | 0.18 |
ENSG00000212572 |
. |
30794 |
0.14 |
| chr4_181237914_181238065 | 0.18 |
NA |
NA |
> 106 |
NA |
| chr3_77047404_77047555 | 0.18 |
ROBO2 |
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
41558 |
0.21 |
| chr6_24926936_24927087 | 0.18 |
FAM65B |
family with sequence similarity 65, member B |
9177 |
0.23 |
| chr17_47084887_47085038 | 0.18 |
RP11-501C14.6 |
|
6125 |
0.1 |
| chr11_14375529_14375805 | 0.17 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
379 |
0.91 |
| chr1_198610531_198610895 | 0.17 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
2421 |
0.36 |
| chr5_112503635_112503786 | 0.17 |
MCC |
mutated in colorectal cancers |
66664 |
0.13 |
| chr11_16761078_16761311 | 0.17 |
C11orf58 |
chromosome 11 open reading frame 58 |
989 |
0.65 |
| chr6_169720130_169720281 | 0.17 |
THBS2 |
thrombospondin 2 |
66066 |
0.13 |
| chr1_242400535_242400686 | 0.17 |
RP11-561I11.3 |
|
33753 |
0.18 |
| chr6_69869443_69869594 | 0.17 |
ENSG00000265498 |
. |
55184 |
0.16 |
| chr1_192776419_192777001 | 0.16 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
1461 |
0.52 |
| chr22_38578075_38578268 | 0.16 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
334 |
0.84 |
| chr6_10310725_10311140 | 0.16 |
ENSG00000221583 |
. |
88663 |
0.07 |
| chr8_49322264_49322415 | 0.16 |
ENSG00000252710 |
. |
101749 |
0.08 |
| chr10_29932434_29932585 | 0.16 |
SVIL |
supervillin |
8608 |
0.22 |
| chr10_91092076_91093023 | 0.16 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
308 |
0.86 |
| chr19_31210066_31211017 | 0.16 |
ENSG00000223148 |
. |
105414 |
0.08 |
| chr13_44310211_44310362 | 0.16 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
50758 |
0.18 |
| chr13_98859522_98859872 | 0.16 |
ENSG00000263399 |
. |
1081 |
0.44 |
| chr12_63025061_63025357 | 0.16 |
ENSG00000238475 |
. |
19746 |
0.14 |
| chr2_216003041_216003192 | 0.16 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
11 |
0.99 |
| chr1_8156287_8156493 | 0.15 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
70022 |
0.1 |
| chrX_98993158_98993309 | 0.15 |
ENSG00000252430 |
. |
352959 |
0.01 |
| chr22_47023615_47023766 | 0.15 |
GRAMD4 |
GRAM domain containing 4 |
1032 |
0.62 |
| chr4_57302350_57302924 | 0.15 |
PAICS |
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
319 |
0.75 |
| chr15_60883034_60883482 | 0.15 |
RORA |
RAR-related orphan receptor A |
1482 |
0.47 |
| chr3_121450033_121450184 | 0.15 |
GOLGB1 |
golgin B1 |
1311 |
0.47 |
| chr19_6464115_6464545 | 0.15 |
CRB3 |
crumbs homolog 3 (Drosophila) |
36 |
0.92 |
| chr8_49621928_49622079 | 0.15 |
EFCAB1 |
EF-hand calcium binding domain 1 |
20345 |
0.28 |
| chr10_5490099_5490572 | 0.15 |
NET1 |
neuroepithelial cell transforming 1 |
1761 |
0.33 |
| chr8_76319339_76319966 | 0.15 |
HNF4G |
hepatocyte nuclear factor 4, gamma |
619 |
0.85 |
| chr6_74512871_74513022 | 0.15 |
RP11-553A21.3 |
|
107092 |
0.06 |
| chr11_134145279_134145568 | 0.15 |
GLB1L3 |
galactosidase, beta 1-like 3 |
1212 |
0.41 |
| chr12_116192167_116192318 | 0.15 |
ENSG00000240205 |
. |
36712 |
0.21 |
| chr7_139047352_139047690 | 0.15 |
C7orf55-LUC7L2 |
C7orf55-LUC7L2 readthrough |
2867 |
0.19 |
| chr11_99364685_99364836 | 0.15 |
CNTN5 |
contactin 5 |
62110 |
0.14 |
| chr4_113331922_113332073 | 0.14 |
RP11-402J6.1 |
|
104544 |
0.06 |
| chr2_177515764_177515915 | 0.14 |
ENSG00000252027 |
. |
13565 |
0.29 |
| chr4_106634812_106634963 | 0.14 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
2884 |
0.28 |
| chr6_137582969_137583120 | 0.14 |
IFNGR1 |
interferon gamma receptor 1 |
42458 |
0.19 |
| chr3_120397291_120397574 | 0.14 |
HGD |
homogentisate 1,2-dioxygenase |
3528 |
0.29 |
| chr11_9272154_9272305 | 0.14 |
RP11-5L12.1 |
|
5173 |
0.21 |
| chr6_16595154_16595305 | 0.14 |
RP1-151F17.1 |
|
166140 |
0.04 |
| chr13_64460857_64461008 | 0.14 |
ENSG00000202478 |
. |
102998 |
0.08 |
| chr10_81841980_81842261 | 0.14 |
TMEM254 |
transmembrane protein 254 |
641 |
0.7 |
| chr14_65170645_65170998 | 0.14 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
1 |
0.98 |
| chr2_61112921_61113072 | 0.14 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
4205 |
0.23 |
| chr6_2952235_2952386 | 0.14 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
9251 |
0.16 |
| chr6_147379941_147380092 | 0.14 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
60583 |
0.16 |
| chr8_119669828_119669979 | 0.14 |
SAMD12 |
sterile alpha motif domain containing 12 |
35669 |
0.24 |
| chr18_27841796_27841947 | 0.13 |
ENSG00000251702 |
. |
722201 |
0.0 |
| chr2_51447556_51447707 | 0.13 |
NRXN1 |
neurexin 1 |
187957 |
0.03 |
| chr1_207413756_207413907 | 0.13 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
81022 |
0.09 |
| chr12_76271082_76271233 | 0.13 |
ENSG00000243420 |
. |
80753 |
0.1 |
| chr16_20774903_20775254 | 0.13 |
ACSM3 |
acyl-CoA synthetase medium-chain family member 3 |
54 |
0.96 |
| chr12_122519567_122519958 | 0.13 |
MLXIP |
MLX interacting protein |
3134 |
0.29 |
| chr10_71529381_71529532 | 0.13 |
COL13A1 |
collagen, type XIII, alpha 1 |
32188 |
0.17 |
| chr2_115486319_115486470 | 0.13 |
DPP10-AS3 |
DPP10 antisense RNA 3 |
104731 |
0.08 |
| chr9_1232780_1232931 | 0.13 |
ENSG00000202383 |
. |
100982 |
0.08 |
| chr21_27936458_27936609 | 0.13 |
CYYR1 |
cysteine/tyrosine-rich 1 |
8962 |
0.3 |
| chrX_57931578_57932092 | 0.13 |
ZXDA |
zinc finger, X-linked, duplicated A |
5232 |
0.36 |
| chr14_21511304_21511455 | 0.13 |
RNASE7 |
ribonuclease, RNase A family, 7 |
994 |
0.3 |
| chr5_67576888_67577103 | 0.13 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
860 |
0.74 |
| chrX_105065279_105065430 | 0.13 |
NRK |
Nik related kinase |
1182 |
0.6 |
| chr3_1166731_1166882 | 0.13 |
CNTN6 |
contactin 6 |
32461 |
0.22 |
| chr6_100722020_100722171 | 0.13 |
RP1-121G13.2 |
|
152899 |
0.04 |
| chr8_133328849_133329000 | 0.13 |
KCNQ3 |
potassium voltage-gated channel, KQT-like subfamily, member 3 |
130585 |
0.05 |
| chr5_95615743_95615894 | 0.12 |
ENSG00000206997 |
. |
69887 |
0.12 |
| chr7_22861355_22861506 | 0.12 |
TOMM7 |
translocase of outer mitochondrial membrane 7 homolog (yeast) |
533 |
0.81 |
| chrX_135436156_135436307 | 0.12 |
GPR112 |
G protein-coupled receptor 112 |
48051 |
0.14 |
| chr5_23013590_23013917 | 0.12 |
CDH12 |
cadherin 12, type 2 (N-cadherin 2) |
160022 |
0.04 |
| chr1_153585227_153585451 | 0.12 |
S100A16 |
S100 calcium binding protein A16 |
220 |
0.83 |
| chr18_61220889_61221040 | 0.12 |
SERPINB12 |
serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
2429 |
0.28 |
| chrX_131405137_131405331 | 0.12 |
RAP2C |
RAP2C, member of RAS oncogene family |
51773 |
0.14 |
| chr6_144585691_144586280 | 0.12 |
UTRN |
utrophin |
20852 |
0.24 |
| chr6_118638895_118639096 | 0.12 |
PLN |
phospholamban |
230466 |
0.02 |
| chr4_124571165_124571642 | 0.12 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
250280 |
0.02 |
| chr3_128892694_128892845 | 0.12 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
9973 |
0.13 |
| chr4_182964416_182964567 | 0.12 |
AC108142.1 |
|
41746 |
0.18 |
| chr18_13019627_13019778 | 0.12 |
CEP192 |
centrosomal protein 192kDa |
10084 |
0.21 |
| chr8_142289533_142290316 | 0.12 |
RP11-10J21.3 |
Uncharacterized protein |
25260 |
0.12 |
| chr2_102466743_102466894 | 0.12 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
5621 |
0.28 |
| chr2_206678700_206678851 | 0.11 |
AC007362.3 |
|
50045 |
0.15 |
| chr8_42356526_42357223 | 0.11 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
1887 |
0.34 |
| chr18_29033854_29034005 | 0.11 |
DSG3 |
desmoglein 3 |
6171 |
0.18 |
| chr17_70583135_70583286 | 0.11 |
ENSG00000200783 |
. |
77081 |
0.11 |
| chr5_67564084_67564251 | 0.11 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
11901 |
0.28 |
| chr2_184151507_184151754 | 0.11 |
NUP35 |
nucleoporin 35kDa |
162457 |
0.04 |
| chr18_4269017_4269168 | 0.11 |
DLGAP1-AS5 |
DLGAP1 antisense RNA 5 |
4459 |
0.35 |
| chr17_12887644_12888019 | 0.11 |
ARHGAP44 |
Rho GTPase activating protein 44 |
4406 |
0.18 |
| chr15_93428286_93428633 | 0.11 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
1933 |
0.32 |
| chr2_182168845_182168996 | 0.11 |
ENSG00000266705 |
. |
1459 |
0.58 |
| chr9_79502275_79502437 | 0.11 |
PRUNE2 |
prune homolog 2 (Drosophila) |
18645 |
0.27 |
| chr17_48800568_48800788 | 0.11 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
3670 |
0.16 |
| chr2_44518711_44518931 | 0.11 |
SLC3A1 |
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
6187 |
0.21 |
| chr6_55730770_55730921 | 0.11 |
BMP5 |
bone morphogenetic protein 5 |
8832 |
0.33 |
| chr6_144809166_144809317 | 0.11 |
UTRN |
utrophin |
95105 |
0.09 |
| chr6_109581664_109581815 | 0.11 |
ENSG00000238474 |
. |
30822 |
0.16 |
| chr10_116285976_116286984 | 0.10 |
ABLIM1 |
actin binding LIM protein 1 |
114 |
0.98 |
| chr1_28696522_28697055 | 0.10 |
PHACTR4 |
phosphatase and actin regulator 4 |
674 |
0.67 |
| chr7_47071998_47072149 | 0.10 |
TNS3 |
tensin 3 |
247843 |
0.02 |
| chr6_136760831_136760982 | 0.10 |
MAP7 |
microtubule-associated protein 7 |
27107 |
0.21 |
| chr11_69969718_69969869 | 0.10 |
RP11-805J14.3 |
|
5610 |
0.17 |
| chr12_62657622_62657800 | 0.10 |
USP15 |
ubiquitin specific peptidase 15 |
3502 |
0.25 |
| chr13_97910664_97910819 | 0.10 |
MBNL2 |
muscleblind-like splicing regulator 2 |
17717 |
0.27 |
| chr2_117398214_117398365 | 0.10 |
ENSG00000239185 |
. |
499062 |
0.0 |
| chr6_142698058_142698209 | 0.10 |
GPR126 |
G protein-coupled receptor 126 |
13316 |
0.29 |
| chr5_154145914_154146065 | 0.10 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
9255 |
0.18 |
| chr18_59208775_59208926 | 0.10 |
CDH20 |
cadherin 20, type 2 |
8249 |
0.32 |
| chr3_142682580_142682815 | 0.10 |
RP11-372E1.6 |
|
203 |
0.7 |
| chr4_43019583_43019734 | 0.10 |
GRXCR1 |
glutaredoxin, cysteine rich 1 |
124374 |
0.06 |
| chr3_171918528_171919229 | 0.10 |
ENSG00000243398 |
. |
36238 |
0.18 |
| chr12_68715253_68715404 | 0.10 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
10488 |
0.26 |
| chr6_10082497_10082648 | 0.10 |
OFCC1 |
orofacial cleft 1 candidate 1 |
32435 |
0.24 |
| chr17_56490822_56490973 | 0.10 |
RNF43 |
ring finger protein 43 |
2105 |
0.25 |
| chr9_7515477_7515628 | 0.10 |
TMEM261 |
transmembrane protein 261 |
284515 |
0.01 |
| chr7_55055085_55055236 | 0.10 |
EGFR |
epidermal growth factor receptor |
31554 |
0.23 |
| chr17_81047529_81047808 | 0.10 |
METRNL |
meteorin, glial cell differentiation regulator-like |
4326 |
0.27 |
| chr6_116381607_116382020 | 0.10 |
FRK |
fyn-related kinase |
108 |
0.97 |
| chr10_102650198_102650475 | 0.10 |
ENSG00000222072 |
. |
16701 |
0.13 |
| chr12_27262305_27262456 | 0.10 |
C12orf71 |
chromosome 12 open reading frame 71 |
26933 |
0.19 |
| chr12_46757806_46758524 | 0.10 |
SLC38A2 |
solute carrier family 38, member 2 |
925 |
0.64 |
| chr3_107317640_107318380 | 0.10 |
BBX |
bobby sox homolog (Drosophila) |
150 |
0.98 |
| chr10_71712113_71712264 | 0.10 |
COL13A1 |
collagen, type XIII, alpha 1 |
21979 |
0.22 |
| chr17_71306920_71307184 | 0.10 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
495 |
0.81 |
| chr1_198418768_198418919 | 0.10 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
90961 |
0.09 |
| chr4_99849016_99849252 | 0.09 |
RP11-571L19.7 |
|
914 |
0.37 |
| chr3_141597606_141597757 | 0.09 |
ATP1B3 |
ATPase, Na+/K+ transporting, beta 3 polypeptide |
1310 |
0.45 |
| chr15_34974520_34974671 | 0.09 |
GJD2 |
gap junction protein, delta 2, 36kDa |
72571 |
0.08 |
| chr15_90543812_90544634 | 0.09 |
ZNF710 |
zinc finger protein 710 |
401 |
0.81 |
| chr5_52546058_52546209 | 0.09 |
MOCS2 |
molybdenum cofactor synthesis 2 |
140240 |
0.04 |
| chr17_62287879_62288139 | 0.09 |
TEX2 |
testis expressed 2 |
2070 |
0.34 |
| chr8_92210223_92210374 | 0.09 |
ENSG00000266194 |
. |
7415 |
0.2 |
| chr5_14205024_14205555 | 0.09 |
TRIO |
trio Rho guanine nucleotide exchange factor |
21382 |
0.29 |
| chr15_51317270_51317421 | 0.09 |
RP11-394B5.2 |
|
44438 |
0.16 |
| chr16_88007697_88007848 | 0.09 |
BANP |
BTG3 associated nuclear protein |
4148 |
0.26 |
| chr8_41181710_41181861 | 0.09 |
SFRP1 |
secreted frizzled-related protein 1 |
14769 |
0.17 |
| chr10_14606601_14606845 | 0.09 |
FAM107B |
family with sequence similarity 107, member B |
7306 |
0.26 |
| chr7_148724822_148725626 | 0.09 |
PDIA4 |
protein disulfide isomerase family A, member 4 |
336 |
0.84 |
| chrX_127442791_127442942 | 0.09 |
ACTRT1 |
actin-related protein T1 |
256484 |
0.02 |
| chr5_34687146_34687427 | 0.09 |
RAI14 |
retinoic acid induced 14 |
378 |
0.9 |
| chr1_68214173_68214559 | 0.09 |
ENSG00000238778 |
. |
23970 |
0.19 |
| chr9_135548642_135548835 | 0.09 |
DDX31 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
2950 |
0.25 |
| chr16_66541951_66542102 | 0.09 |
TK2 |
thymidine kinase 2, mitochondrial |
5660 |
0.12 |
| chr21_42441242_42441393 | 0.09 |
LINC00323 |
long intergenic non-protein coding RNA 323 |
78033 |
0.09 |
| chr1_222440358_222440509 | 0.08 |
ENSG00000222399 |
. |
236644 |
0.02 |
| chr13_49081946_49082097 | 0.08 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
25192 |
0.23 |
| chr10_121412015_121412434 | 0.08 |
BAG3 |
BCL2-associated athanogene 3 |
1342 |
0.5 |
| chr3_175025439_175025620 | 0.08 |
NAALADL2-AS2 |
NAALADL2 antisense RNA 2 |
36644 |
0.19 |
| chr3_161089901_161090298 | 0.08 |
SPTSSB |
serine palmitoyltransferase, small subunit B |
569 |
0.86 |
| chr15_71006570_71006722 | 0.08 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
11032 |
0.26 |
| chr2_165776815_165776966 | 0.08 |
SLC38A11 |
solute carrier family 38, member 11 |
17043 |
0.17 |
| chr1_33447039_33447276 | 0.08 |
RP1-117O3.2 |
|
5519 |
0.15 |
| chrX_133523066_133523217 | 0.08 |
ENSG00000251962 |
. |
14271 |
0.17 |
| chrX_119678777_119678928 | 0.08 |
ENSG00000272179 |
. |
682 |
0.7 |
| chr18_63063126_63063277 | 0.08 |
CDH7 |
cadherin 7, type 2 |
354287 |
0.01 |
| chr15_94782960_94783136 | 0.08 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
8281 |
0.33 |
| chr3_190313233_190313384 | 0.08 |
IL1RAP |
interleukin 1 receptor accessory protein |
19789 |
0.23 |
| chr18_12255048_12255199 | 0.08 |
CIDEA |
cell death-inducing DFFA-like effector a |
805 |
0.66 |
| chr18_29952348_29952499 | 0.08 |
RP11-344B2.2 |
|
85023 |
0.08 |
| chr1_244212846_244213916 | 0.08 |
ZBTB18 |
zinc finger and BTB domain containing 18 |
1204 |
0.52 |
| chr2_113538908_113539059 | 0.08 |
IL1A |
interleukin 1, alpha |
3184 |
0.22 |
| chr18_26683920_26684071 | 0.08 |
ENSG00000265730 |
. |
8352 |
0.28 |
| chr19_4970283_4970930 | 0.08 |
KDM4B |
lysine (K)-specific demethylase 4B |
1463 |
0.39 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.0 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.0 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |