Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
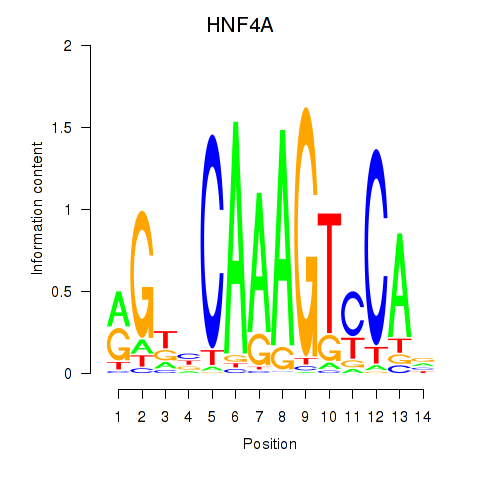
Results for HNF4A
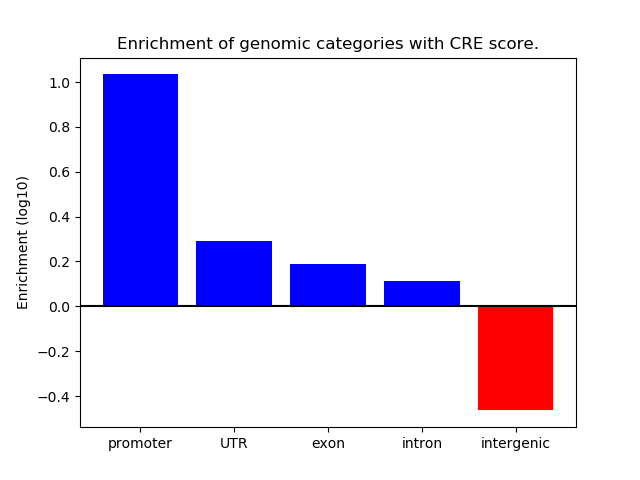
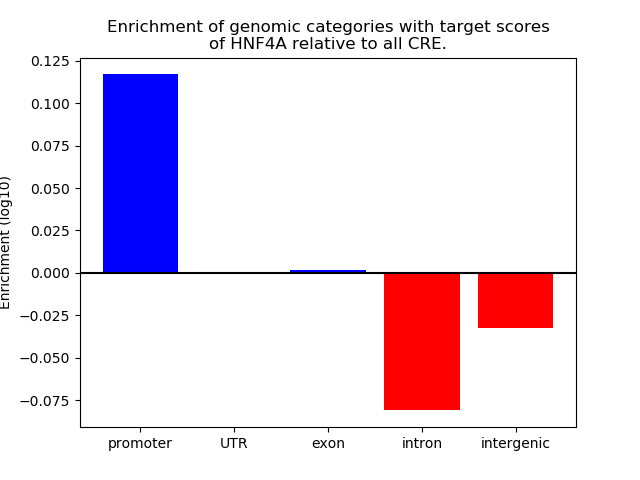
Z-value: 0.52
Transcription factors associated with HNF4A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF4A
|
ENSG00000101076.12 | hepatocyte nuclear factor 4 alpha |
Activity of the HNF4A motif across conditions
Conditions sorted by the z-value of the HNF4A motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
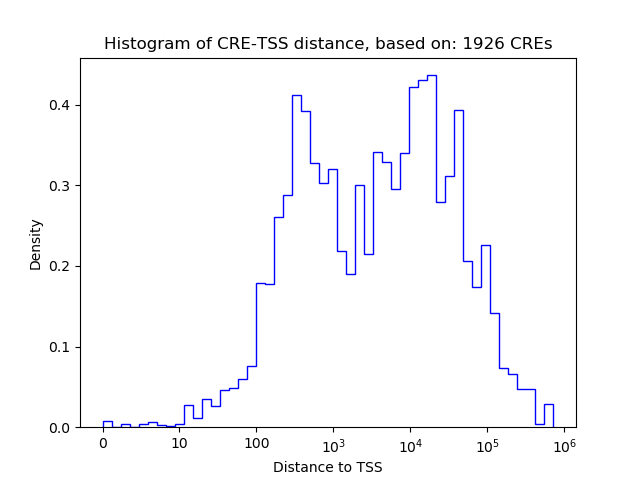
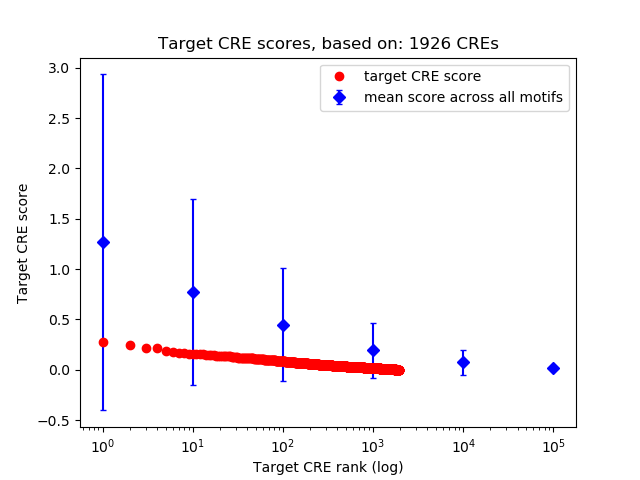
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_121077248_121077399 | 0.28 |
COL14A1 |
collagen, type XIV, alpha 1 |
40493 |
0.2 |
| chr11_63535609_63536120 | 0.24 |
ENSG00000264519 |
. |
29679 |
0.13 |
| chr2_26634976_26635146 | 0.22 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
10277 |
0.21 |
| chr1_27890528_27890679 | 0.21 |
RP1-159A19.4 |
|
38287 |
0.11 |
| chr1_40402828_40403049 | 0.19 |
ENSG00000207356 |
. |
7732 |
0.14 |
| chr2_198573276_198573488 | 0.18 |
MARS2 |
methionyl-tRNA synthetase 2, mitochondrial |
3295 |
0.23 |
| chr17_4615541_4615774 | 0.17 |
ARRB2 |
arrestin, beta 2 |
1663 |
0.18 |
| chr8_30085369_30085520 | 0.17 |
RP11-51J9.4 |
|
47040 |
0.14 |
| chr19_54494957_54495171 | 0.16 |
CACNG6 |
calcium channel, voltage-dependent, gamma subunit 6 |
478 |
0.67 |
| chr12_3861008_3861159 | 0.16 |
EFCAB4B |
EF-hand calcium binding domain 4B |
1180 |
0.56 |
| chr2_223920421_223920654 | 0.16 |
KCNE4 |
potassium voltage-gated channel, Isk-related family, member 4 |
3675 |
0.34 |
| chr20_25176247_25177242 | 0.16 |
ENTPD6 |
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
269 |
0.91 |
| chr14_105531321_105531563 | 0.16 |
GPR132 |
G protein-coupled receptor 132 |
325 |
0.89 |
| chr1_145454279_145454446 | 0.15 |
TXNIP |
thioredoxin interacting protein |
15056 |
0.1 |
| chr2_47039434_47039760 | 0.15 |
LINC01118 |
long intergenic non-protein coding RNA 1118 |
4225 |
0.19 |
| chr1_203254106_203254257 | 0.14 |
BTG2 |
BTG family, member 2 |
20483 |
0.15 |
| chr7_37956036_37956797 | 0.14 |
SFRP4 |
secreted frizzled-related protein 4 |
101 |
0.97 |
| chr16_16213974_16214125 | 0.14 |
RP11-517A5.7 |
|
102835 |
0.05 |
| chr2_74729539_74729700 | 0.14 |
LBX2-AS1 |
LBX2 antisense RNA 1 |
103 |
0.77 |
| chr20_11916577_11916728 | 0.14 |
BTBD3 |
BTB (POZ) domain containing 3 |
17153 |
0.23 |
| chrX_123094531_123095374 | 0.14 |
STAG2 |
stromal antigen 2 |
212 |
0.95 |
| chr22_37257050_37257437 | 0.14 |
NCF4 |
neutrophil cytosolic factor 4, 40kDa |
213 |
0.91 |
| chr1_200200915_200201066 | 0.14 |
ENSG00000221403 |
. |
87028 |
0.09 |
| chr1_31255877_31256028 | 0.14 |
LAPTM5 |
lysosomal protein transmembrane 5 |
25285 |
0.14 |
| chr21_39756340_39756646 | 0.13 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
87645 |
0.09 |
| chr17_73305250_73305401 | 0.13 |
GRB2 |
growth factor receptor-bound protein 2 |
12509 |
0.09 |
| chr1_198898259_198898514 | 0.13 |
ENSG00000207759 |
. |
70104 |
0.12 |
| chr20_5717977_5718128 | 0.13 |
C20orf196 |
chromosome 20 open reading frame 196 |
12987 |
0.26 |
| chr8_50822371_50822736 | 0.13 |
SNTG1 |
syntrophin, gamma 1 |
204 |
0.97 |
| chr19_2394888_2395039 | 0.13 |
TMPRSS9 |
transmembrane protease, serine 9 |
5179 |
0.14 |
| chr2_135004109_135004260 | 0.13 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
7646 |
0.25 |
| chr3_3214293_3214763 | 0.12 |
CRBN |
cereblon |
6830 |
0.19 |
| chr6_99292512_99292663 | 0.12 |
POU3F2 |
POU class 3 homeobox 2 |
10007 |
0.31 |
| chr7_7091088_7091239 | 0.12 |
ENSG00000264679 |
. |
15160 |
0.1 |
| chr7_38385599_38385750 | 0.12 |
AMPH |
amphiphysin |
117039 |
0.06 |
| chr17_80817564_80818630 | 0.12 |
TBCD |
tubulin folding cofactor D |
173 |
0.94 |
| chr8_2569726_2569923 | 0.12 |
MYOM2 |
myomesin 2 |
576640 |
0.0 |
| chr18_3074243_3074394 | 0.12 |
ENSG00000252353 |
. |
19248 |
0.18 |
| chr8_15869012_15869163 | 0.12 |
MSR1 |
macrophage scavenger receptor 1 |
152673 |
0.05 |
| chr12_132345455_132345606 | 0.12 |
MMP17 |
matrix metallopeptidase 17 (membrane-inserted) |
15690 |
0.16 |
| chr10_129679566_129679717 | 0.12 |
CLRN3 |
clarin 3 |
11570 |
0.22 |
| chr11_67417906_67418392 | 0.12 |
ACY3 |
aspartoacylase (aminocyclase) 3 |
19 |
0.96 |
| chr8_58424780_58424931 | 0.12 |
ENSG00000252057 |
. |
44289 |
0.21 |
| chr15_38855839_38856970 | 0.12 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
432 |
0.84 |
| chr4_40208849_40209185 | 0.11 |
RHOH |
ras homolog family member H |
7053 |
0.23 |
| chr16_73266288_73266597 | 0.11 |
C16orf47 |
chromosome 16 open reading frame 47 |
88096 |
0.1 |
| chr22_46545178_46545717 | 0.11 |
PPARA |
peroxisome proliferator-activated receptor alpha |
977 |
0.45 |
| chr2_162762054_162762205 | 0.11 |
ENSG00000253046 |
. |
13163 |
0.28 |
| chr7_150212054_150212338 | 0.11 |
GIMAP7 |
GTPase, IMAP family member 7 |
278 |
0.91 |
| chr1_211526625_211526879 | 0.11 |
TRAF5 |
TNF receptor-associated factor 5 |
7046 |
0.27 |
| chr2_61023542_61023744 | 0.11 |
PAPOLG |
poly(A) polymerase gamma |
14533 |
0.2 |
| chr7_30543935_30544434 | 0.11 |
GGCT |
gamma-glutamylcyclotransferase |
231 |
0.93 |
| chr22_37260110_37260261 | 0.11 |
NCF4 |
neutrophil cytosolic factor 4, 40kDa |
3155 |
0.18 |
| chr4_39666570_39666736 | 0.11 |
SMIM14 |
small integral membrane protein 14 |
25943 |
0.14 |
| chr2_231508912_231509063 | 0.11 |
ENSG00000199791 |
. |
57151 |
0.11 |
| chr2_113932280_113932569 | 0.11 |
AC016683.5 |
|
490 |
0.59 |
| chr14_39571616_39572168 | 0.11 |
SEC23A |
Sec23 homolog A (S. cerevisiae) |
387 |
0.86 |
| chr6_1885704_1885932 | 0.10 |
FOXC1 |
forkhead box C1 |
275137 |
0.01 |
| chr14_90148063_90148287 | 0.10 |
ENSG00000200312 |
. |
30680 |
0.14 |
| chr20_1855464_1855750 | 0.10 |
SIRPA |
signal-regulatory protein alpha |
19547 |
0.22 |
| chr14_22564190_22564341 | 0.10 |
ENSG00000238634 |
. |
46622 |
0.18 |
| chr8_23396205_23396870 | 0.10 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
9964 |
0.16 |
| chr1_246740599_246740764 | 0.10 |
CNST |
consortin, connexin sorting protein |
10865 |
0.17 |
| chr2_158296405_158296556 | 0.10 |
CYTIP |
cytohesin 1 interacting protein |
554 |
0.73 |
| chr22_35726479_35726752 | 0.10 |
ENSG00000266320 |
. |
5018 |
0.18 |
| chr21_16135398_16135558 | 0.10 |
AF165138.7 |
Protein LOC388813 |
104336 |
0.07 |
| chr7_143083684_143083835 | 0.10 |
ZYX |
zyxin |
3666 |
0.13 |
| chr9_100615486_100616611 | 0.10 |
FOXE1 |
forkhead box E1 (thyroid transcription factor 2) |
512 |
0.81 |
| chr12_121734766_121735534 | 0.10 |
CAMKK2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
11 |
0.98 |
| chr10_90789554_90789771 | 0.10 |
RP11-399O19.9 |
|
14069 |
0.14 |
| chrX_48660892_48661351 | 0.10 |
HDAC6 |
histone deacetylase 6 |
164 |
0.91 |
| chr21_36258327_36258638 | 0.10 |
RUNX1 |
runt-related transcription factor 1 |
998 |
0.7 |
| chr11_18639490_18639676 | 0.10 |
SPTY2D1 |
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
16457 |
0.14 |
| chr13_94514457_94514608 | 0.10 |
GPC6-AS2 |
GPC6 antisense RNA 2 |
26135 |
0.27 |
| chr2_191044944_191045899 | 0.10 |
C2orf88 |
chromosome 2 open reading frame 88 |
168 |
0.96 |
| chr10_65027523_65028177 | 0.09 |
JMJD1C |
jumonji domain containing 1C |
976 |
0.68 |
| chr14_24897194_24897345 | 0.09 |
KHNYN |
KH and NYN domain containing |
1223 |
0.26 |
| chr10_121487526_121487707 | 0.09 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
2007 |
0.37 |
| chr6_43772269_43772420 | 0.09 |
VEGFA |
vascular endothelial growth factor A |
30254 |
0.15 |
| chr7_155025143_155025294 | 0.09 |
AC099552.4 |
Uncharacterized protein |
35072 |
0.16 |
| chr12_1922066_1922217 | 0.09 |
CACNA2D4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
1184 |
0.5 |
| chr6_169295722_169295873 | 0.09 |
SMOC2 |
SPARC related modular calcium binding 2 |
242033 |
0.02 |
| chr10_99496936_99497805 | 0.09 |
ZFYVE27 |
zinc finger, FYVE domain containing 27 |
473 |
0.78 |
| chr13_94834592_94834743 | 0.09 |
GPC6-AS1 |
GPC6 antisense RNA 1 |
5578 |
0.34 |
| chr11_75474128_75474279 | 0.09 |
DGAT2 |
diacylglycerol O-acyltransferase 2 |
3646 |
0.13 |
| chr4_130265390_130265541 | 0.09 |
C4orf33 |
chromosome 4 open reading frame 33 |
248140 |
0.02 |
| chr15_45478994_45479145 | 0.09 |
SHF |
Src homology 2 domain containing F |
1081 |
0.34 |
| chr11_117821640_117821911 | 0.09 |
TMPRSS13 |
transmembrane protease, serine 13 |
21601 |
0.15 |
| chr14_61645817_61645968 | 0.09 |
PRKCH |
protein kinase C, eta |
8385 |
0.26 |
| chr19_29479808_29479959 | 0.09 |
ENSG00000252272 |
. |
12694 |
0.27 |
| chr15_76603307_76603813 | 0.09 |
ETFA |
electron-transfer-flavoprotein, alpha polypeptide |
177 |
0.95 |
| chr22_22215698_22215849 | 0.09 |
MAPK1 |
mitogen-activated protein kinase 1 |
5957 |
0.17 |
| chr1_210411744_210411895 | 0.09 |
SERTAD4-AS1 |
SERTAD4 antisense RNA 1 |
4427 |
0.25 |
| chr11_67777777_67778022 | 0.09 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
109 |
0.94 |
| chr7_38398847_38399253 | 0.09 |
AMPH |
amphiphysin |
103663 |
0.08 |
| chr2_211035606_211035857 | 0.09 |
KANSL1L |
KAT8 regulatory NSL complex subunit 1-like |
320 |
0.44 |
| chrX_154012446_154012633 | 0.09 |
ENSG00000206693 |
. |
9266 |
0.12 |
| chr8_128903916_128904067 | 0.08 |
TMEM75 |
transmembrane protein 75 |
56600 |
0.12 |
| chr2_175547809_175548354 | 0.08 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
454 |
0.87 |
| chr7_105517318_105518128 | 0.08 |
ATXN7L1 |
ataxin 7-like 1 |
673 |
0.81 |
| chr11_65585009_65585678 | 0.08 |
SNX32 |
sorting nexin 32 |
15769 |
0.08 |
| chr5_131132977_131133425 | 0.08 |
FNIP1 |
folliculin interacting protein 1 |
491 |
0.49 |
| chr1_226924448_226924715 | 0.08 |
ITPKB |
inositol-trisphosphate 3-kinase B |
578 |
0.8 |
| chr11_48055572_48055723 | 0.08 |
AC103828.1 |
|
18240 |
0.2 |
| chr10_13571820_13572051 | 0.08 |
BEND7 |
BEN domain containing 7 |
1402 |
0.3 |
| chr3_48935289_48935440 | 0.08 |
SLC25A20 |
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
961 |
0.43 |
| chr14_72020560_72020711 | 0.08 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
24593 |
0.25 |
| chr13_42033808_42033959 | 0.08 |
RGCC |
regulator of cell cycle |
2188 |
0.25 |
| chr1_121260341_121261384 | 0.08 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
324925 |
0.01 |
| chr11_46721727_46722111 | 0.08 |
ARHGAP1 |
Rho GTPase activating protein 1 |
230 |
0.69 |
| chr8_104198051_104198202 | 0.08 |
RP11-318M2.2 |
|
13758 |
0.14 |
| chr1_45274565_45275349 | 0.08 |
BTBD19 |
BTB (POZ) domain containing 19 |
762 |
0.37 |
| chr9_136580274_136580425 | 0.08 |
SARDH |
sarcosine dehydrogenase |
11526 |
0.2 |
| chr1_200422603_200422754 | 0.08 |
ZNF281 |
zinc finger protein 281 |
43494 |
0.2 |
| chr8_42054040_42054280 | 0.08 |
PLAT |
plasminogen activator, tissue |
944 |
0.52 |
| chr12_57442830_57442981 | 0.08 |
MYO1A |
myosin IA |
996 |
0.37 |
| chr17_37041846_37041997 | 0.08 |
LASP1 |
LIM and SH3 protein 1 |
11778 |
0.09 |
| chr1_26348142_26348482 | 0.08 |
EXTL1 |
exostosin-like glycosyltransferase 1 |
41 |
0.95 |
| chr1_207094900_207095227 | 0.08 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
149 |
0.94 |
| chr17_7123160_7124223 | 0.08 |
ACADVL |
acyl-CoA dehydrogenase, very long chain |
424 |
0.48 |
| chr22_43389803_43390178 | 0.08 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
8456 |
0.21 |
| chr19_55834048_55834199 | 0.08 |
TMEM150B |
transmembrane protein 150B |
2304 |
0.12 |
| chrX_119444733_119445129 | 0.08 |
TMEM255A |
transmembrane protein 255A |
357 |
0.89 |
| chr1_182162992_182163143 | 0.08 |
ZNF648 |
zinc finger protein 648 |
132220 |
0.05 |
| chr11_64764535_64764747 | 0.08 |
BATF2 |
basic leucine zipper transcription factor, ATF-like 2 |
124 |
0.92 |
| chr11_92028446_92028637 | 0.08 |
FAT3 |
FAT atypical cadherin 3 |
56721 |
0.17 |
| chr2_43154619_43154770 | 0.08 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
134962 |
0.05 |
| chr12_114744833_114744984 | 0.08 |
TBX5 |
T-box 5 |
96795 |
0.08 |
| chr11_47269852_47270330 | 0.08 |
ACP2 |
acid phosphatase 2, lysosomal |
88 |
0.8 |
| chr10_11637184_11637335 | 0.08 |
RP11-138I18.1 |
|
16045 |
0.22 |
| chr19_15362877_15363312 | 0.07 |
EPHX3 |
epoxide hydrolase 3 |
18848 |
0.15 |
| chr21_45285452_45286244 | 0.07 |
AGPAT3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
781 |
0.65 |
| chr16_3136460_3136756 | 0.07 |
ENSG00000200204 |
. |
464 |
0.56 |
| chr1_211849013_211849381 | 0.07 |
RP11-122M14.1 |
|
93 |
0.7 |
| chr5_36242698_36242954 | 0.07 |
NADK2 |
NAD kinase 2, mitochondrial |
445 |
0.83 |
| chr2_111227830_111228011 | 0.07 |
LIMS3 |
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein |
2473 |
0.24 |
| chr1_156719567_156719969 | 0.07 |
HDGF |
hepatoma-derived growth factor |
1317 |
0.26 |
| chr4_140763091_140763242 | 0.07 |
ENSG00000252233 |
. |
47579 |
0.15 |
| chr19_13213099_13213611 | 0.07 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
326 |
0.79 |
| chr19_6738037_6738395 | 0.07 |
TRIP10 |
thyroid hormone receptor interactor 10 |
280 |
0.7 |
| chr12_118796178_118796667 | 0.07 |
TAOK3 |
TAO kinase 3 |
488 |
0.85 |
| chr8_29630934_29631085 | 0.07 |
ENSG00000221003 |
. |
155112 |
0.04 |
| chr14_23777494_23777674 | 0.07 |
BCL2L2 |
BCL2-like 2 |
434 |
0.58 |
| chr16_81500466_81500617 | 0.07 |
CMIP |
c-Maf inducing protein |
21766 |
0.23 |
| chr17_36119345_36119496 | 0.07 |
HNF1B |
HNF1 homeobox B |
14183 |
0.18 |
| chr2_43202085_43202236 | 0.07 |
ENSG00000207087 |
. |
116472 |
0.06 |
| chr9_35071347_35071984 | 0.07 |
VCP |
valosin containing protein |
346 |
0.77 |
| chrX_2809831_2809982 | 0.07 |
ARSD-AS1 |
ARSD antisense RNA 1 |
13039 |
0.15 |
| chr3_63953694_63954206 | 0.07 |
ATXN7 |
ataxin 7 |
530 |
0.74 |
| chr10_62190052_62190231 | 0.07 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
40653 |
0.22 |
| chr15_49148078_49148229 | 0.07 |
SHC4 |
SHC (Src homology 2 domain containing) family, member 4 |
21533 |
0.14 |
| chr2_242549180_242549748 | 0.07 |
THAP4 |
THAP domain containing 4 |
7441 |
0.13 |
| chr7_129546585_129546878 | 0.07 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
44436 |
0.1 |
| chrX_23802856_23803288 | 0.07 |
SAT1 |
spermidine/spermine N1-acetyltransferase 1 |
1782 |
0.23 |
| chr2_219646650_219647418 | 0.07 |
CYP27A1 |
cytochrome P450, family 27, subfamily A, polypeptide 1 |
555 |
0.7 |
| chrX_65235295_65235677 | 0.07 |
ENSG00000207939 |
. |
3226 |
0.26 |
| chr22_30819496_30819934 | 0.07 |
MTFP1 |
mitochondrial fission process 1 |
1803 |
0.13 |
| chr18_19809259_19809410 | 0.07 |
RP11-627G18.1 |
|
24142 |
0.15 |
| chr3_123601954_123602216 | 0.07 |
MYLK |
myosin light chain kinase |
1064 |
0.59 |
| chr18_60536807_60536958 | 0.07 |
AC015989.2 |
|
30934 |
0.17 |
| chr6_15421960_15422111 | 0.07 |
JARID2 |
jumonji, AT rich interactive domain 2 |
20946 |
0.24 |
| chr3_119421432_119421650 | 0.07 |
MAATS1 |
MYCBP-associated, testis expressed 1 |
328 |
0.87 |
| chr8_1940163_1940314 | 0.07 |
KBTBD11 |
kelch repeat and BTB (POZ) domain containing 11 |
18194 |
0.24 |
| chr6_33377620_33378073 | 0.07 |
PHF1 |
PHD finger protein 1 |
330 |
0.79 |
| chr19_48866469_48867161 | 0.07 |
TMEM143 |
transmembrane protein 143 |
116 |
0.89 |
| chr2_110658685_110658836 | 0.07 |
LIMS3 |
LIM and senescent cell antigen-like domains 3 |
2492 |
0.39 |
| chr11_63370704_63370855 | 0.07 |
PLA2G16 |
phospholipase A2, group XVI |
5274 |
0.15 |
| chr1_163044093_163044377 | 0.07 |
RGS4 |
regulator of G-protein signaling 4 |
2299 |
0.4 |
| chr1_203273544_203273695 | 0.07 |
BTG2 |
BTG family, member 2 |
1045 |
0.51 |
| chr8_99422129_99422280 | 0.07 |
KCNS2 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
17046 |
0.19 |
| chr8_41998685_41998836 | 0.07 |
RP11-589C21.5 |
|
11521 |
0.17 |
| chr2_232410055_232410615 | 0.07 |
NMUR1 |
neuromedin U receptor 1 |
15129 |
0.14 |
| chr5_94405852_94406003 | 0.07 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
11259 |
0.29 |
| chr19_10416066_10416280 | 0.07 |
ZGLP1 |
zinc finger, GATA-like protein 1 |
4286 |
0.08 |
| chr5_154026662_154027279 | 0.06 |
ENSG00000221552 |
. |
38366 |
0.13 |
| chr11_78127359_78128498 | 0.06 |
GAB2 |
GRB2-associated binding protein 2 |
897 |
0.59 |
| chr12_89551363_89551514 | 0.06 |
ENSG00000238302 |
. |
124624 |
0.06 |
| chr1_113615415_113615661 | 0.06 |
LRIG2 |
leucine-rich repeats and immunoglobulin-like domains 2 |
293 |
0.94 |
| chr2_181012083_181012234 | 0.06 |
CWC22 |
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
140318 |
0.05 |
| chr16_89003788_89003939 | 0.06 |
RP11-830F9.7 |
|
301 |
0.83 |
| chr1_198131206_198131357 | 0.06 |
NEK7 |
NIMA-related kinase 7 |
5030 |
0.35 |
| chr5_32444599_32444828 | 0.06 |
ZFR |
zinc finger RNA binding protein |
154 |
0.96 |
| chr5_159375097_159375248 | 0.06 |
ADRA1B |
adrenoceptor alpha 1B |
31382 |
0.18 |
| chr21_36412378_36412763 | 0.06 |
RUNX1 |
runt-related transcription factor 1 |
8892 |
0.33 |
| chr11_78021681_78021861 | 0.06 |
RP11-452H21.1 |
|
14033 |
0.18 |
| chr14_65439795_65439946 | 0.06 |
RAB15 |
RAB15, member RAS oncogene family |
376 |
0.79 |
| chr13_34110879_34111030 | 0.06 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
186187 |
0.03 |
| chr17_33868045_33868636 | 0.06 |
SLFN12L |
schlafen family member 12-like |
3460 |
0.14 |
| chr17_27942716_27943482 | 0.06 |
CORO6 |
coronin 6 |
2036 |
0.14 |
| chr12_9768605_9768756 | 0.06 |
ENSG00000212345 |
. |
8144 |
0.13 |
| chr1_33463602_33463869 | 0.06 |
RP1-117O3.2 |
|
11059 |
0.14 |
| chr8_38795554_38795750 | 0.06 |
CTD-2544N14.3 |
|
34878 |
0.11 |
| chr19_10197336_10197761 | 0.06 |
C19orf66 |
chromosome 19 open reading frame 66 |
20 |
0.94 |
| chr5_73936545_73937281 | 0.06 |
ENC1 |
ectodermal-neural cortex 1 (with BTB domain) |
336 |
0.81 |
| chr22_26062428_26062638 | 0.06 |
ENSG00000222585 |
. |
48603 |
0.13 |
| chr6_109330916_109331330 | 0.06 |
SESN1 |
sestrin 1 |
365 |
0.87 |
| chr20_30457693_30458808 | 0.06 |
DUSP15 |
dual specificity phosphatase 15 |
125 |
0.67 |
| chr10_122708557_122709094 | 0.06 |
RP11-95I16.4 |
|
68646 |
0.12 |
| chr8_97657180_97658239 | 0.06 |
CPQ |
carboxypeptidase Q |
106 |
0.98 |
| chr7_23513291_23513443 | 0.06 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
3281 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:1903313 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.0 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |