Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
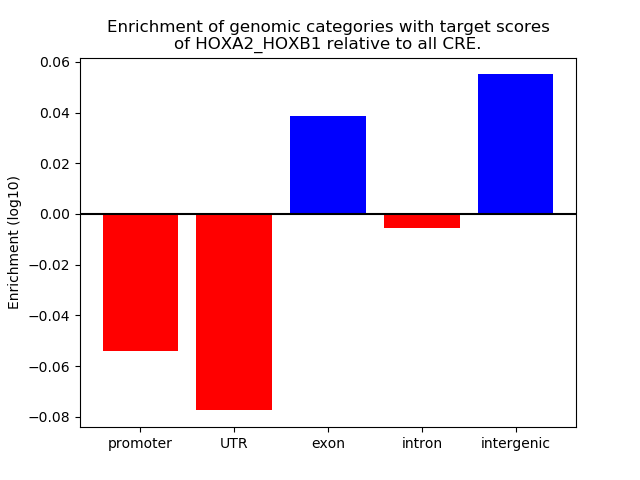
Results for HOXA2_HOXB1
Z-value: 0.40
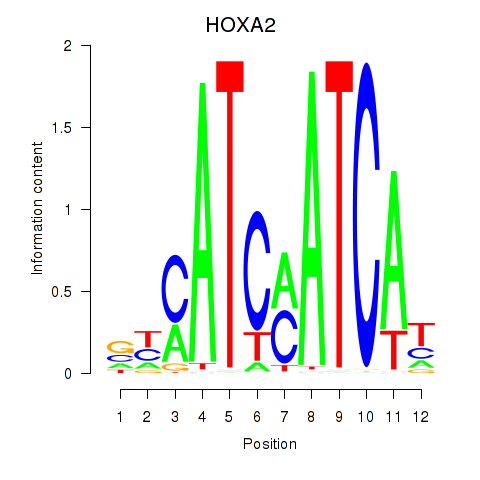
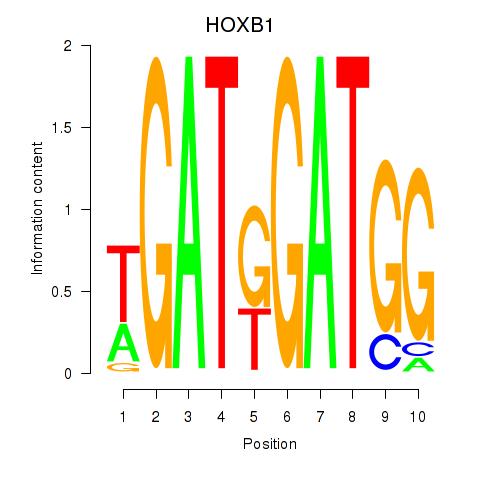
Transcription factors associated with HOXA2_HOXB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA2
|
ENSG00000105996.5 | homeobox A2 |
|
HOXB1
|
ENSG00000120094.6 | homeobox B1 |
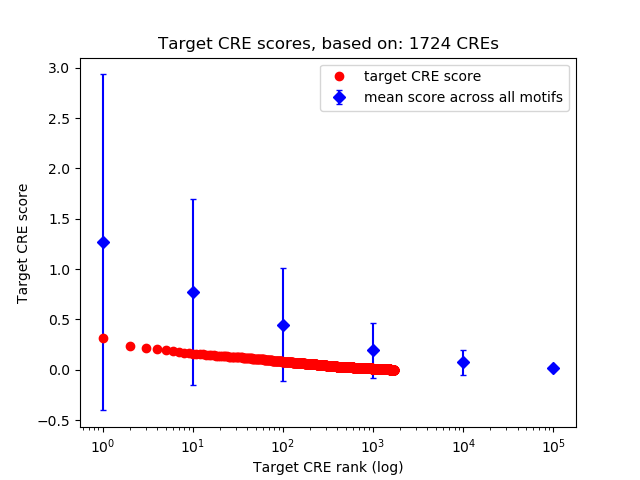
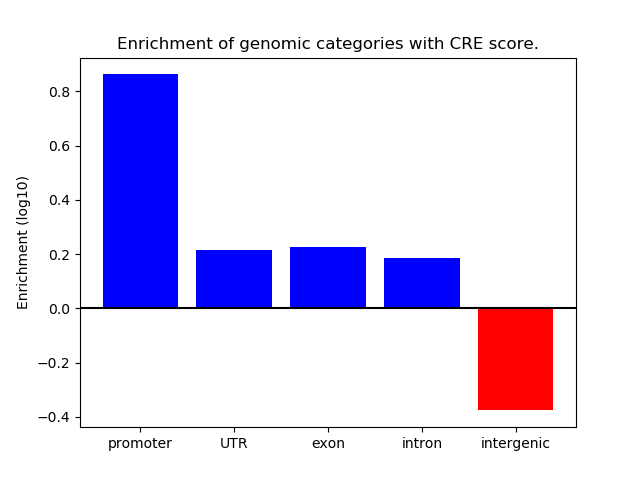
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_27142124_27142281 | HOXA2 | 228 | 0.820740 | 0.84 | 4.9e-03 | Click! |
| chr7_27142737_27142888 | HOXA2 | 382 | 0.674861 | 0.84 | 5.1e-03 | Click! |
| chr7_27140714_27141098 | HOXA2 | 1524 | 0.169156 | 0.81 | 7.5e-03 | Click! |
| chr7_27143124_27143275 | HOXA2 | 769 | 0.384045 | 0.77 | 1.6e-02 | Click! |
| chr7_27142394_27142575 | HOXA2 | 54 | 0.930137 | 0.74 | 2.2e-02 | Click! |
| chr17_46606887_46607038 | HOXB1 | 1310 | 0.246061 | 0.74 | 2.3e-02 | Click! |
| chr17_46607108_46607259 | HOXB1 | 1089 | 0.300034 | 0.68 | 4.3e-02 | Click! |
Activity of the HOXA2_HOXB1 motif across conditions
Conditions sorted by the z-value of the HOXA2_HOXB1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_121494510_121494857 | 0.31 |
GLI2 |
GLI family zinc finger 2 |
860 |
0.74 |
| chr9_34745263_34745414 | 0.24 |
FAM205A |
family with sequence similarity 205, member A |
15874 |
0.13 |
| chr17_17317253_17317688 | 0.22 |
ENSG00000201741 |
. |
46288 |
0.12 |
| chr12_85305887_85306488 | 0.21 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
384 |
0.92 |
| chr11_122072235_122072431 | 0.20 |
ENSG00000207994 |
. |
49317 |
0.12 |
| chr5_100168975_100169126 | 0.19 |
ENSG00000221263 |
. |
16781 |
0.26 |
| chr19_464760_464911 | 0.18 |
SHC2 |
SHC (Src homology 2 domain containing) transforming protein 2 |
3839 |
0.12 |
| chr16_16115866_16116017 | 0.17 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
12228 |
0.24 |
| chr3_159482295_159482916 | 0.16 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
287 |
0.9 |
| chr2_53971971_53972122 | 0.16 |
GPR75-ASB3 |
GPR75-ASB3 readthrough |
20640 |
0.16 |
| chr11_34706995_34707273 | 0.16 |
EHF |
ets homologous factor |
42960 |
0.19 |
| chrX_45482024_45482567 | 0.15 |
ENSG00000207870 |
. |
123399 |
0.06 |
| chr2_54684400_54684738 | 0.15 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
171 |
0.97 |
| chr1_83285095_83285395 | 0.15 |
LPHN2 |
latrophilin 2 |
839672 |
0.0 |
| chr14_53158961_53159226 | 0.15 |
ERO1L |
ERO1-like (S. cerevisiae) |
3154 |
0.22 |
| chr1_87244034_87244185 | 0.15 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
73531 |
0.09 |
| chr2_74968036_74968578 | 0.14 |
SEMA4F |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
86869 |
0.07 |
| chr7_22233477_22233744 | 0.14 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
83 |
0.98 |
| chr15_74880905_74881056 | 0.14 |
CLK3 |
CDC-like kinase 3 |
19733 |
0.14 |
| chr18_53214040_53214242 | 0.14 |
TCF4 |
transcription factor 4 |
36141 |
0.18 |
| chr5_10757151_10757327 | 0.14 |
CTD-2154B17.4 |
|
3938 |
0.26 |
| chr14_103588344_103588755 | 0.13 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
1249 |
0.43 |
| chr1_208137959_208138110 | 0.13 |
CD34 |
CD34 molecule |
53287 |
0.17 |
| chr21_43422426_43422615 | 0.13 |
ZBTB21 |
zinc finger and BTB domain containing 21 |
5646 |
0.21 |
| chr7_5467738_5468320 | 0.13 |
TNRC18 |
trinucleotide repeat containing 18 |
2984 |
0.19 |
| chr20_61468182_61468333 | 0.13 |
COL9A3 |
collagen, type IX, alpha 3 |
19094 |
0.11 |
| chr11_96062035_96062186 | 0.13 |
ENSG00000266192 |
. |
12492 |
0.2 |
| chr2_222367084_222367382 | 0.13 |
EPHA4 |
EPH receptor A4 |
47 |
0.98 |
| chr4_8702215_8702366 | 0.13 |
CPZ |
carboxypeptidase Z |
98593 |
0.07 |
| chr13_30009560_30009804 | 0.13 |
MTUS2 |
microtubule associated tumor suppressor candidate 2 |
6836 |
0.28 |
| chr7_100773540_100774051 | 0.13 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
3416 |
0.13 |
| chr11_27739578_27739815 | 0.13 |
BDNF |
brain-derived neurotrophic factor |
1598 |
0.49 |
| chr17_36623721_36623872 | 0.13 |
ARHGAP23 |
Rho GTPase activating protein 23 |
4384 |
0.19 |
| chrX_130902480_130902797 | 0.12 |
ENSG00000200587 |
. |
170346 |
0.03 |
| chr9_27408498_27408649 | 0.12 |
RP11-298E2.2 |
|
16841 |
0.25 |
| chr21_28215601_28215791 | 0.12 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
163 |
0.97 |
| chr10_105614245_105615135 | 0.12 |
SH3PXD2A |
SH3 and PX domains 2A |
474 |
0.82 |
| chr18_74798828_74799607 | 0.12 |
MBP |
myelin basic protein |
18000 |
0.27 |
| chr10_28491840_28491991 | 0.12 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
35751 |
0.21 |
| chr14_81893853_81894004 | 0.12 |
STON2 |
stonin 2 |
180 |
0.97 |
| chr2_183225036_183225187 | 0.11 |
ENSG00000242121 |
. |
46100 |
0.17 |
| chr11_30606067_30606222 | 0.11 |
RP5-1024C24.1 |
|
434 |
0.82 |
| chr14_50468177_50468632 | 0.11 |
C14orf182 |
chromosome 14 open reading frame 182 |
5834 |
0.2 |
| chr16_5648936_5649087 | 0.11 |
RP11-124K4.1 |
|
12283 |
0.3 |
| chr8_25793197_25793466 | 0.11 |
EBF2 |
early B-cell factor 2 |
47899 |
0.19 |
| chr14_24019702_24019853 | 0.11 |
ZFHX2 |
zinc finger homeobox 2 |
1081 |
0.33 |
| chr3_156480231_156480382 | 0.11 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
63783 |
0.12 |
| chr11_3181150_3181578 | 0.11 |
OSBPL5 |
oxysterol binding protein-like 5 |
1259 |
0.4 |
| chr19_10736478_10736985 | 0.11 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
530 |
0.65 |
| chr18_9107793_9107944 | 0.11 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
3788 |
0.2 |
| chr8_22250007_22250158 | 0.11 |
SLC39A14 |
solute carrier family 39 (zinc transporter), member 14 |
261 |
0.91 |
| chr7_75924108_75924402 | 0.11 |
ENSG00000263569 |
. |
6147 |
0.16 |
| chr12_132629597_132629942 | 0.11 |
NOC4L |
nucleolar complex associated 4 homolog (S. cerevisiae) |
460 |
0.65 |
| chr6_102178821_102179006 | 0.11 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
54269 |
0.19 |
| chr7_156851742_156851893 | 0.11 |
MNX1-AS1 |
MNX1 antisense RNA 1 (head to head) |
48318 |
0.11 |
| chr10_33620428_33620906 | 0.11 |
NRP1 |
neuropilin 1 |
2643 |
0.37 |
| chr2_134391396_134391547 | 0.10 |
ENSG00000200708 |
. |
37809 |
0.2 |
| chr16_49890203_49890582 | 0.10 |
ZNF423 |
zinc finger protein 423 |
346 |
0.91 |
| chrX_11456470_11456766 | 0.10 |
ARHGAP6 |
Rho GTPase activating protein 6 |
10725 |
0.3 |
| chr17_71068251_71068402 | 0.10 |
SLC39A11 |
solute carrier family 39, member 11 |
17577 |
0.24 |
| chr15_95869891_95870263 | 0.10 |
ENSG00000222076 |
. |
418956 |
0.01 |
| chr15_32976738_32976906 | 0.10 |
RP11-1000B6.2 |
|
11556 |
0.16 |
| chr18_26092204_26092423 | 0.10 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
334903 |
0.01 |
| chr2_28178102_28178322 | 0.10 |
ENSG00000265321 |
. |
41022 |
0.15 |
| chr8_37378833_37379079 | 0.10 |
RP11-150O12.6 |
|
4417 |
0.31 |
| chr10_110004648_110004799 | 0.10 |
ENSG00000222436 |
. |
696345 |
0.0 |
| chr10_5670766_5671061 | 0.10 |
ENSG00000240577 |
. |
2513 |
0.27 |
| chr3_98619314_98619465 | 0.10 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
626 |
0.66 |
| chr7_24576451_24576602 | 0.10 |
ENSG00000206877 |
. |
425 |
0.89 |
| chr17_39577204_39577355 | 0.10 |
KRT37 |
keratin 37 |
3496 |
0.11 |
| chr5_159738704_159739455 | 0.09 |
CCNJL |
cyclin J-like |
404 |
0.84 |
| chr3_129204924_129205100 | 0.09 |
IFT122 |
intraflagellar transport 122 homolog (Chlamydomonas) |
2022 |
0.27 |
| chr3_159451541_159451741 | 0.09 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
29861 |
0.17 |
| chr2_180990617_180990780 | 0.09 |
CWC22 |
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
118858 |
0.07 |
| chr7_123389310_123389704 | 0.09 |
RP11-390E23.6 |
|
385 |
0.47 |
| chr9_86892548_86892699 | 0.09 |
RP11-380F14.2 |
|
511 |
0.86 |
| chr17_38267528_38268059 | 0.09 |
MSL1 |
male-specific lethal 1 homolog (Drosophila) |
10758 |
0.11 |
| chr17_69972427_69972645 | 0.09 |
AC007461.1 |
Uncharacterized protein |
63628 |
0.15 |
| chr14_55137368_55137519 | 0.09 |
SAMD4A |
sterile alpha motif domain containing 4A |
84108 |
0.09 |
| chr8_26370931_26371439 | 0.09 |
PNMA2 |
paraneoplastic Ma antigen 2 |
423 |
0.6 |
| chr21_33244856_33245438 | 0.09 |
HUNK |
hormonally up-regulated Neu-associated kinase |
481 |
0.88 |
| chr2_65804604_65804761 | 0.09 |
ENSG00000251900 |
. |
22014 |
0.21 |
| chr7_23509552_23510001 | 0.09 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
310 |
0.88 |
| chr7_131582308_131582459 | 0.09 |
ENSG00000252849 |
. |
18698 |
0.25 |
| chr7_44922681_44922832 | 0.09 |
ENSG00000264326 |
. |
1357 |
0.26 |
| chr3_72131377_72131562 | 0.09 |
ENSG00000212070 |
. |
180110 |
0.03 |
| chr8_59956242_59956393 | 0.09 |
RP11-328K2.1 |
|
50820 |
0.14 |
| chr12_114844132_114844283 | 0.09 |
TBX5 |
T-box 5 |
318 |
0.89 |
| chr8_116463616_116464117 | 0.09 |
TRPS1 |
trichorhinophalangeal syndrome I |
40582 |
0.2 |
| chr3_13689202_13689353 | 0.09 |
LINC00620 |
long intergenic non-protein coding RNA 620 |
2919 |
0.28 |
| chr7_25891701_25892366 | 0.09 |
ENSG00000199085 |
. |
97573 |
0.09 |
| chr10_8201743_8201894 | 0.09 |
GATA3 |
GATA binding protein 3 |
105049 |
0.08 |
| chr10_77168132_77169350 | 0.09 |
ENSG00000237149 |
. |
5929 |
0.2 |
| chr9_128512876_128513197 | 0.09 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
2558 |
0.37 |
| chr10_127779801_127780118 | 0.08 |
ENSG00000222740 |
. |
54192 |
0.14 |
| chr2_205180048_205180265 | 0.08 |
PARD3B |
par-3 family cell polarity regulator beta |
230360 |
0.02 |
| chr5_15501266_15501663 | 0.08 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
83 |
0.99 |
| chr2_114035017_114035267 | 0.08 |
PAX8 |
paired box 8 |
829 |
0.59 |
| chr17_75471247_75471398 | 0.08 |
SEPT9 |
septin 9 |
3 |
0.97 |
| chr4_160220043_160220341 | 0.08 |
ENSG00000216089 |
. |
3326 |
0.3 |
| chr2_42796417_42796822 | 0.08 |
MTA3 |
metastasis associated 1 family, member 3 |
95 |
0.98 |
| chr15_37394480_37395011 | 0.08 |
MEIS2 |
Meis homeobox 2 |
1241 |
0.49 |
| chr3_61580182_61580471 | 0.08 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
32741 |
0.25 |
| chr12_47671859_47672010 | 0.08 |
RP11-493L12.3 |
|
870 |
0.66 |
| chr3_68697737_68697888 | 0.08 |
ENSG00000212095 |
. |
13745 |
0.3 |
| chr12_25877270_25877421 | 0.08 |
ENSG00000222950 |
. |
55158 |
0.14 |
| chr11_61780107_61780258 | 0.08 |
AP003733.1 |
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
44729 |
0.1 |
| chr2_27301899_27302732 | 0.08 |
EMILIN1 |
elastin microfibril interfacer 1 |
880 |
0.32 |
| chr12_54672473_54672704 | 0.08 |
CBX5 |
chromobox homolog 5 |
1298 |
0.19 |
| chr7_93485635_93485786 | 0.08 |
TFPI2 |
tissue factor pathway inhibitor 2 |
33772 |
0.15 |
| chr5_134722208_134722661 | 0.08 |
H2AFY |
H2A histone family, member Y |
12467 |
0.16 |
| chr12_123574141_123574292 | 0.08 |
PITPNM2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
8336 |
0.19 |
| chr4_6992958_6993109 | 0.08 |
AC097382.5 |
|
4035 |
0.13 |
| chr3_78853664_78853815 | 0.08 |
ENSG00000240964 |
. |
133703 |
0.05 |
| chr2_235403849_235404784 | 0.08 |
ARL4C |
ADP-ribosylation factor-like 4C |
928 |
0.74 |
| chr1_8202972_8203123 | 0.08 |
ENSG00000200975 |
. |
63610 |
0.11 |
| chr11_122028064_122028812 | 0.08 |
ENSG00000207994 |
. |
5422 |
0.19 |
| chr9_89069847_89070053 | 0.08 |
ENSG00000222293 |
. |
31907 |
0.23 |
| chr4_105747663_105747814 | 0.08 |
ENSG00000251906 |
. |
148081 |
0.04 |
| chr3_61549164_61549336 | 0.08 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
1665 |
0.55 |
| chr10_99932028_99932445 | 0.08 |
R3HCC1L |
R3H domain and coiled-coil containing 1-like |
9596 |
0.25 |
| chr16_85645006_85645418 | 0.08 |
GSE1 |
Gse1 coiled-coil protein |
197 |
0.95 |
| chr1_165868486_165868637 | 0.07 |
RP11-525G13.2 |
|
1359 |
0.44 |
| chrX_106984928_106985096 | 0.07 |
TSC22D3 |
TSC22 domain family, member 3 |
19510 |
0.2 |
| chr1_56547076_56547335 | 0.07 |
PIGQP1 |
phosphatidylinositol glycan anchor biosynthesis, class Q pseudogene 1 |
142252 |
0.05 |
| chr10_102322629_102322843 | 0.07 |
HIF1AN |
hypoxia inducible factor 1, alpha subunit inhibitor |
27120 |
0.15 |
| chr2_14896967_14897186 | 0.07 |
AC011897.1 |
Uncharacterized protein |
121861 |
0.06 |
| chr9_685775_685926 | 0.07 |
RP11-130C19.3 |
|
295 |
0.92 |
| chr14_21093852_21094122 | 0.07 |
OR6S1 |
olfactory receptor, family 6, subfamily S, member 1 |
15863 |
0.1 |
| chr1_84084018_84084169 | 0.07 |
ENSG00000223231 |
. |
175467 |
0.03 |
| chr15_33148553_33148741 | 0.07 |
FMN1 |
formin 1 |
31808 |
0.17 |
| chr13_98826337_98826632 | 0.07 |
RNF113B |
ring finger protein 113B |
3035 |
0.21 |
| chr5_59508772_59508923 | 0.07 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
27422 |
0.24 |
| chr21_42213476_42213627 | 0.07 |
DSCAM |
Down syndrome cell adhesion molecule |
5514 |
0.33 |
| chr19_5681315_5681466 | 0.07 |
RPL36 |
ribosomal protein L36 |
197 |
0.45 |
| chr5_17021245_17021396 | 0.07 |
BASP1 |
brain abundant, membrane attached signal protein 1 |
44387 |
0.15 |
| chr14_88921501_88921652 | 0.07 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
14384 |
0.17 |
| chr4_172734546_172734879 | 0.07 |
GALNTL6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
58 |
0.99 |
| chr3_169377883_169378034 | 0.07 |
MECOM |
MDS1 and EVI1 complex locus |
3220 |
0.23 |
| chr10_63734627_63734778 | 0.07 |
ENSG00000221272 |
. |
47925 |
0.17 |
| chr2_138723355_138723506 | 0.07 |
HNMT |
histamine N-methyltransferase |
1387 |
0.61 |
| chr5_14265991_14266171 | 0.07 |
TRIO |
trio Rho guanine nucleotide exchange factor |
25005 |
0.28 |
| chr16_88228496_88228647 | 0.07 |
BANP |
BTG3 associated nuclear protein |
224947 |
0.02 |
| chr3_98612768_98613177 | 0.07 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
7043 |
0.19 |
| chr6_142335140_142335592 | 0.07 |
RP11-137J7.2 |
|
74004 |
0.11 |
| chr1_87239567_87240110 | 0.07 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
69260 |
0.09 |
| chr6_33384595_33384833 | 0.07 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
984 |
0.34 |
| chr7_83823215_83823431 | 0.07 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
894 |
0.76 |
| chr18_45968069_45968289 | 0.07 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
31056 |
0.17 |
| chr1_54793079_54793243 | 0.07 |
RP5-997D24.3 |
|
42083 |
0.14 |
| chr1_21650807_21651093 | 0.07 |
ECE1 |
endothelin converting enzyme 1 |
21047 |
0.18 |
| chr2_31636273_31636424 | 0.07 |
XDH |
xanthine dehydrogenase |
1233 |
0.61 |
| chr10_94581983_94582410 | 0.07 |
EXOC6 |
exocyst complex component 6 |
8739 |
0.28 |
| chr16_65566073_65566543 | 0.07 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
410040 |
0.01 |
| chr16_51465176_51465516 | 0.07 |
ENSG00000223168 |
. |
240128 |
0.02 |
| chr3_52140725_52140876 | 0.07 |
LINC00696 |
long intergenic non-protein coding RNA 696 |
43233 |
0.09 |
| chr4_102267340_102267537 | 0.07 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
515 |
0.73 |
| chr6_163664912_163665063 | 0.07 |
ENSG00000239136 |
. |
17045 |
0.25 |
| chr4_26203423_26203574 | 0.07 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
38421 |
0.23 |
| chr3_42697942_42698093 | 0.07 |
RP4-613B23.1 |
|
2137 |
0.17 |
| chr4_159732250_159732401 | 0.07 |
FNIP2 |
folliculin interacting protein 2 |
4925 |
0.23 |
| chr18_61148002_61148153 | 0.06 |
SERPINB5 |
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
3717 |
0.21 |
| chr8_135790645_135790802 | 0.06 |
ENSG00000207582 |
. |
22127 |
0.21 |
| chrX_123128269_123128420 | 0.06 |
STAG2 |
stromal antigen 2 |
31324 |
0.2 |
| chr4_143739251_143739537 | 0.06 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
28049 |
0.27 |
| chr18_4269017_4269168 | 0.06 |
DLGAP1-AS5 |
DLGAP1 antisense RNA 5 |
4459 |
0.35 |
| chr11_12699867_12700018 | 0.06 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
2892 |
0.38 |
| chr19_914980_915131 | 0.06 |
R3HDM4 |
R3H domain containing 4 |
1815 |
0.15 |
| chr2_12458046_12458293 | 0.06 |
ENSG00000207183 |
. |
93458 |
0.09 |
| chr15_72529055_72529849 | 0.06 |
PKM |
pyruvate kinase, muscle |
5288 |
0.17 |
| chr12_21927150_21927652 | 0.06 |
KCNJ8 |
potassium inwardly-rectifying channel, subfamily J, member 8 |
354 |
0.86 |
| chr4_157862974_157863267 | 0.06 |
PDGFC |
platelet derived growth factor C |
28935 |
0.19 |
| chr2_100635044_100635528 | 0.06 |
AFF3 |
AF4/FMR2 family, member 3 |
85706 |
0.09 |
| chr2_85640026_85640421 | 0.06 |
CAPG |
capping protein (actin filament), gelsolin-like |
939 |
0.34 |
| chr7_6309088_6309239 | 0.06 |
CYTH3 |
cytohesin 3 |
3112 |
0.25 |
| chr8_131144625_131144776 | 0.06 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
13932 |
0.2 |
| chr1_164535324_164535475 | 0.06 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
3357 |
0.34 |
| chr5_142625895_142626046 | 0.06 |
ARHGAP26 |
Rho GTPase activating protein 26 |
39205 |
0.19 |
| chr6_121756849_121757212 | 0.06 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
192 |
0.94 |
| chr7_2235658_2235809 | 0.06 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
27530 |
0.17 |
| chr10_53222210_53222361 | 0.06 |
RP11-539E19.2 |
|
159959 |
0.04 |
| chr4_184827832_184828262 | 0.06 |
STOX2 |
storkhead box 2 |
1538 |
0.51 |
| chr17_4711009_4711212 | 0.06 |
PLD2 |
phospholipase D2 |
253 |
0.82 |
| chr7_16168928_16169137 | 0.06 |
ISPD-AS1 |
ISPD antisense RNA 1 |
81081 |
0.11 |
| chr7_17315289_17315760 | 0.06 |
AC003075.4 |
|
5403 |
0.27 |
| chr10_11872880_11873031 | 0.06 |
PROSER2 |
proline and serine-rich protein 2 |
727 |
0.72 |
| chr14_21731452_21731695 | 0.06 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
737 |
0.64 |
| chr14_62232461_62232666 | 0.06 |
SNAPC1 |
small nuclear RNA activating complex, polypeptide 1, 43kDa |
3488 |
0.33 |
| chr10_119312526_119312804 | 0.06 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
8086 |
0.24 |
| chr6_164616974_164617125 | 0.06 |
ENSG00000266128 |
. |
355456 |
0.01 |
| chr6_168719854_168720005 | 0.06 |
DACT2 |
dishevelled-binding antagonist of beta-catenin 2 |
461 |
0.86 |
| chr6_76458966_76459788 | 0.06 |
MYO6 |
myosin VI |
328 |
0.89 |
| chr7_39641818_39642080 | 0.06 |
AC004837.4 |
|
7940 |
0.18 |
| chr17_62655006_62655429 | 0.06 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
2813 |
0.3 |
| chr10_102505217_102505492 | 0.06 |
PAX2 |
paired box 2 |
114 |
0.98 |
| chr22_46460492_46460792 | 0.06 |
RP6-109B7.4 |
|
5129 |
0.11 |
| chr16_66752038_66752189 | 0.06 |
RP11-63M22.2 |
|
2687 |
0.2 |
| chr11_119583124_119583275 | 0.06 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
16064 |
0.18 |
| chr1_26496798_26497060 | 0.06 |
RP11-96L14.7 |
|
375 |
0.52 |
| chr6_8434475_8434734 | 0.06 |
SLC35B3 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
1112 |
0.69 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.1 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:1901889 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |