Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
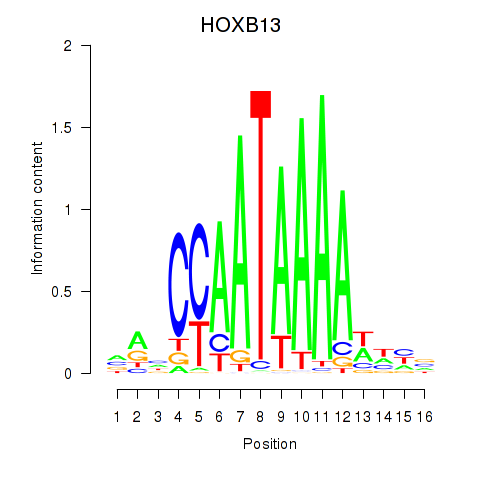
Results for HOXB13
Z-value: 0.92
Transcription factors associated with HOXB13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB13
|
ENSG00000159184.7 | homeobox B13 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_46805922_46806163 | HOXB13 | 498 | 0.644655 | 0.10 | 7.9e-01 | Click! |
Activity of the HOXB13 motif across conditions
Conditions sorted by the z-value of the HOXB13 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
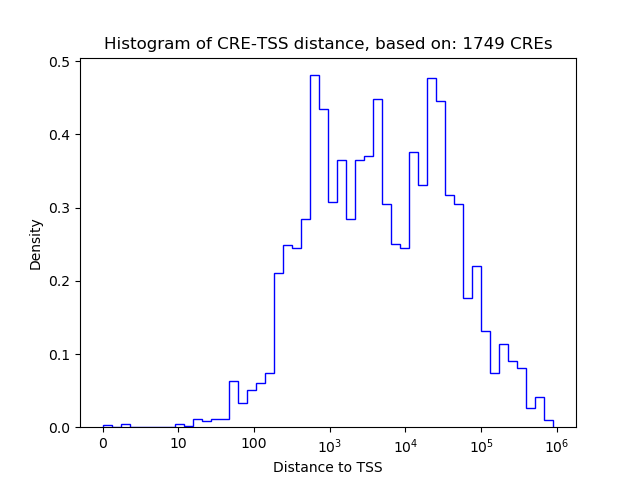
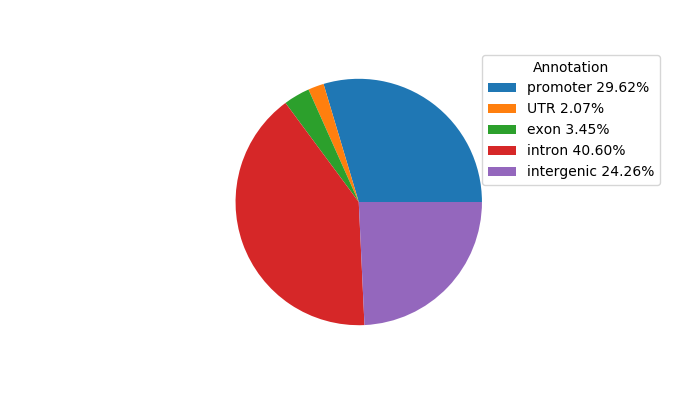
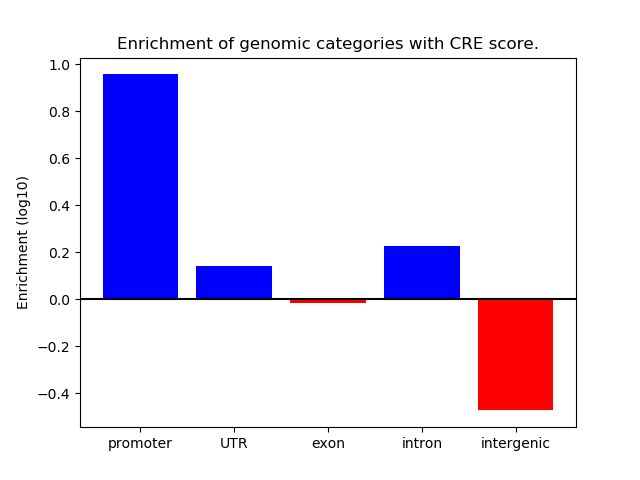
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_109086470_109087224 | 0.66 |
LEF1 |
lymphoid enhancer-binding factor 1 |
610 |
0.72 |
| chr3_151964474_151964662 | 0.53 |
MBNL1 |
muscleblind-like splicing regulator 1 |
21261 |
0.21 |
| chr1_248729795_248729949 | 0.45 |
RP11-438F14.3 |
|
3971 |
0.12 |
| chr3_63954713_63955178 | 0.45 |
ATXN7 |
ataxin 7 |
1525 |
0.34 |
| chr11_5246641_5246814 | 0.43 |
ENSG00000221031 |
. |
858 |
0.32 |
| chr13_110441094_110441245 | 0.43 |
IRS2 |
insulin receptor substrate 2 |
2254 |
0.42 |
| chr5_80257520_80257735 | 0.43 |
CTC-459I6.1 |
|
901 |
0.48 |
| chr2_173305484_173305639 | 0.43 |
AC078883.4 |
|
12230 |
0.17 |
| chr14_22974481_22974956 | 0.42 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
18547 |
0.09 |
| chr1_167458198_167458349 | 0.42 |
CD247 |
CD247 molecule |
29502 |
0.16 |
| chr2_143889174_143889389 | 0.40 |
ARHGAP15 |
Rho GTPase activating protein 15 |
2398 |
0.38 |
| chr1_22402292_22402667 | 0.39 |
CDC42-IT1 |
CDC42 intronic transcript 1 (non-protein coding) |
16789 |
0.13 |
| chr9_100682755_100682906 | 0.39 |
C9orf156 |
chromosome 9 open reading frame 156 |
1939 |
0.27 |
| chr13_41589998_41590808 | 0.39 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
3047 |
0.26 |
| chr9_4285477_4285628 | 0.38 |
GLIS3 |
GLIS family zinc finger 3 |
12944 |
0.21 |
| chr4_39033572_39034012 | 0.36 |
TMEM156 |
transmembrane protein 156 |
249 |
0.93 |
| chr2_106412175_106412698 | 0.36 |
NCK2 |
NCK adaptor protein 2 |
20579 |
0.25 |
| chr16_1430217_1430368 | 0.35 |
UNKL |
unkempt family zinc finger-like |
605 |
0.41 |
| chr3_59996849_59997073 | 0.35 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
39378 |
0.24 |
| chrX_78200973_78201449 | 0.35 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
293 |
0.95 |
| chr6_109196078_109196273 | 0.35 |
ARMC2 |
armadillo repeat containing 2 |
5994 |
0.26 |
| chr4_154412529_154412680 | 0.33 |
KIAA0922 |
KIAA0922 |
25103 |
0.22 |
| chr7_19955546_19955697 | 0.33 |
AC005062.2 |
|
52239 |
0.16 |
| chr4_39660337_39660488 | 0.32 |
SMIM14 |
small integral membrane protein 14 |
19702 |
0.16 |
| chr4_26858665_26858816 | 0.31 |
STIM2 |
stromal interaction molecule 2 |
560 |
0.8 |
| chr14_102278079_102278230 | 0.30 |
CTD-2017C7.2 |
|
1496 |
0.3 |
| chr4_40210765_40210916 | 0.30 |
RHOH |
ras homolog family member H |
8876 |
0.22 |
| chr15_81586633_81587120 | 0.30 |
IL16 |
interleukin 16 |
2378 |
0.31 |
| chr2_87830138_87830289 | 0.30 |
RP11-1399P15.1 |
|
52660 |
0.16 |
| chr21_34697768_34697948 | 0.29 |
IFNAR1 |
interferon (alpha, beta and omega) receptor 1 |
580 |
0.74 |
| chr12_25206187_25206540 | 0.29 |
LRMP |
lymphoid-restricted membrane protein |
689 |
0.71 |
| chr7_35730534_35730808 | 0.29 |
HERPUD2 |
HERPUD family member 2 |
3505 |
0.27 |
| chr7_136976771_136976922 | 0.29 |
ENSG00000202023 |
. |
4412 |
0.31 |
| chr17_46506355_46506700 | 0.29 |
SKAP1 |
src kinase associated phosphoprotein 1 |
1025 |
0.46 |
| chr2_111616545_111616696 | 0.28 |
ACOXL |
acyl-CoA oxidase-like |
53724 |
0.17 |
| chr5_150460010_150460252 | 0.27 |
TNIP1 |
TNFAIP3 interacting protein 1 |
446 |
0.84 |
| chr18_60985812_60986160 | 0.27 |
BCL2 |
B-cell CLL/lymphoma 2 |
59 |
0.97 |
| chr2_112177530_112177803 | 0.27 |
ENSG00000266139 |
. |
98998 |
0.09 |
| chr20_10600368_10600683 | 0.27 |
JAG1 |
jagged 1 |
42629 |
0.18 |
| chr19_42056498_42056882 | 0.27 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
804 |
0.6 |
| chr1_198620599_198620750 | 0.27 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
12382 |
0.23 |
| chr10_121487824_121487975 | 0.27 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
2290 |
0.34 |
| chr4_39032194_39032374 | 0.27 |
TMEM156 |
transmembrane protein 156 |
1757 |
0.38 |
| chr16_81748402_81748687 | 0.26 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
24158 |
0.24 |
| chr16_87825792_87825943 | 0.26 |
KLHDC4 |
kelch domain containing 4 |
26285 |
0.14 |
| chr5_14039928_14040178 | 0.26 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
95401 |
0.09 |
| chr1_112167264_112167415 | 0.26 |
RAP1A |
RAP1A, member of RAS oncogene family |
4570 |
0.2 |
| chr6_27099516_27099912 | 0.26 |
HIST1H2BJ |
histone cluster 1, H2bj |
613 |
0.5 |
| chr2_68379959_68380110 | 0.26 |
WDR92 |
WD repeat domain 92 |
4571 |
0.18 |
| chr11_5713127_5713492 | 0.25 |
TRIM22 |
tripartite motif containing 22 |
1052 |
0.38 |
| chr16_9189012_9189240 | 0.24 |
C16orf72 |
chromosome 16 open reading frame 72 |
3621 |
0.19 |
| chr17_74656206_74656421 | 0.24 |
ENSG00000199735 |
. |
3911 |
0.11 |
| chr18_9150083_9150234 | 0.24 |
ANKRD12 |
ankyrin repeat domain 12 |
12597 |
0.16 |
| chr3_98610682_98610833 | 0.24 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
9258 |
0.18 |
| chr13_109783688_109783839 | 0.24 |
MYO16-AS2 |
MYO16 antisense RNA 2 |
29496 |
0.24 |
| chr12_96042633_96042784 | 0.23 |
ENSG00000212448 |
. |
31243 |
0.19 |
| chr9_128586328_128586530 | 0.23 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
41121 |
0.2 |
| chr6_53174830_53174981 | 0.23 |
ENSG00000264056 |
. |
33114 |
0.15 |
| chr6_21109076_21109227 | 0.22 |
ENSG00000252046 |
. |
115352 |
0.07 |
| chr17_64818195_64818346 | 0.22 |
CACNG5 |
calcium channel, voltage-dependent, gamma subunit 5 |
12965 |
0.2 |
| chr10_23634424_23634575 | 0.22 |
C10orf67 |
chromosome 10 open reading frame 67 |
725 |
0.59 |
| chr18_28567286_28567437 | 0.21 |
ENSG00000251702 |
. |
3289 |
0.27 |
| chr11_128894218_128894369 | 0.21 |
ARHGAP32 |
Rho GTPase activating protein 32 |
205 |
0.94 |
| chr1_111212136_111212287 | 0.21 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
5444 |
0.2 |
| chr4_148982006_148982383 | 0.21 |
RP11-76G10.1 |
|
85428 |
0.1 |
| chr4_129732983_129734245 | 0.21 |
JADE1 |
jade family PHD finger 1 |
615 |
0.83 |
| chr17_19702847_19702998 | 0.21 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
3482 |
0.23 |
| chr10_105670292_105670878 | 0.21 |
OBFC1 |
oligonucleotide/oligosaccharide-binding fold containing 1 |
6842 |
0.18 |
| chr11_104715007_104715158 | 0.20 |
CASP12 |
caspase 12 (gene/pseudogene) |
54059 |
0.14 |
| chr2_99345755_99346000 | 0.20 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
834 |
0.68 |
| chr2_160817314_160817486 | 0.20 |
LY75-CD302 |
LY75-CD302 readthrough |
56179 |
0.13 |
| chr18_2662663_2662814 | 0.20 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
6852 |
0.17 |
| chr6_44214898_44215120 | 0.20 |
HSP90AB1 |
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
185 |
0.9 |
| chr15_94040362_94040513 | 0.20 |
ENSG00000212063 |
. |
210885 |
0.02 |
| chr21_20920825_20920976 | 0.20 |
ENSG00000212609 |
. |
203435 |
0.03 |
| chr20_52446261_52446776 | 0.19 |
AC005220.3 |
|
110181 |
0.06 |
| chr14_102283767_102283918 | 0.19 |
CTD-2017C7.2 |
|
7184 |
0.15 |
| chr10_22938432_22938680 | 0.19 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
57914 |
0.15 |
| chr13_48984563_48985023 | 0.19 |
LPAR6 |
lysophosphatidic acid receptor 6 |
16250 |
0.26 |
| chr3_60061066_60061217 | 0.19 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
103558 |
0.09 |
| chr2_237446083_237446280 | 0.19 |
IQCA1 |
IQ motif containing with AAA domain 1 |
29996 |
0.19 |
| chr16_84766426_84766761 | 0.19 |
USP10 |
ubiquitin specific peptidase 10 |
32947 |
0.16 |
| chr21_25801249_25801648 | 0.19 |
ENSG00000232512 |
. |
671997 |
0.0 |
| chr20_54993887_54994066 | 0.19 |
CASS4 |
Cas scaffolding protein family member 4 |
6659 |
0.14 |
| chr13_99953472_99953749 | 0.19 |
GPR183 |
G protein-coupled receptor 183 |
6049 |
0.22 |
| chr11_67009621_67010037 | 0.19 |
KDM2A |
lysine (K)-specific demethylase 2A |
2311 |
0.2 |
| chr1_26453918_26454263 | 0.18 |
PDIK1L |
PDLIM1 interacting kinase 1 like |
15750 |
0.1 |
| chr14_22466666_22466817 | 0.18 |
ENSG00000238634 |
. |
144146 |
0.04 |
| chr6_27833598_27833859 | 0.18 |
HIST1H2AL |
histone cluster 1, H2al |
694 |
0.38 |
| chr10_74008100_74008285 | 0.18 |
DDIT4 |
DNA-damage-inducible transcript 4 |
25486 |
0.12 |
| chr2_198169992_198170200 | 0.18 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
2853 |
0.22 |
| chr11_59383655_59383910 | 0.18 |
AP000442.1 |
|
120 |
0.56 |
| chr6_34283507_34283709 | 0.18 |
C6orf1 |
chromosome 6 open reading frame 1 |
66361 |
0.09 |
| chr12_9103508_9103764 | 0.18 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
996 |
0.36 |
| chr12_43354342_43354493 | 0.17 |
ENSG00000215993 |
. |
330966 |
0.01 |
| chrX_135861825_135862156 | 0.17 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
2257 |
0.28 |
| chr13_74159314_74159465 | 0.17 |
ENSG00000206697 |
. |
358105 |
0.01 |
| chr13_114240659_114240810 | 0.17 |
TFDP1 |
transcription factor Dp-1 |
993 |
0.58 |
| chr3_31575511_31576107 | 0.17 |
STT3B |
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
1527 |
0.55 |
| chr5_40683307_40683699 | 0.17 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
3903 |
0.25 |
| chr15_60841858_60842044 | 0.17 |
CTD-2501E16.2 |
|
19779 |
0.18 |
| chr15_31631556_31631837 | 0.17 |
KLF13 |
Kruppel-like factor 13 |
440 |
0.9 |
| chr12_104858566_104858717 | 0.17 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
7862 |
0.28 |
| chr5_79514748_79514899 | 0.16 |
ENSG00000239159 |
. |
20585 |
0.18 |
| chr20_3137491_3137642 | 0.16 |
UBOX5 |
U-box domain containing 5 |
2956 |
0.15 |
| chr20_31988765_31988916 | 0.16 |
CDK5RAP1 |
CDK5 regulatory subunit associated protein 1 |
483 |
0.8 |
| chr14_22984271_22984422 | 0.16 |
TRAJ15 |
T cell receptor alpha joining 15 |
14234 |
0.1 |
| chr1_236960314_236960595 | 0.16 |
MTR |
5-methyltetrahydrofolate-homocysteine methyltransferase |
1472 |
0.52 |
| chr2_197024603_197024962 | 0.16 |
STK17B |
serine/threonine kinase 17b |
3411 |
0.25 |
| chr6_144163877_144164309 | 0.16 |
LTV1 |
LTV1 homolog (S. cerevisiae) |
388 |
0.89 |
| chr7_41921705_41921953 | 0.16 |
AC005027.3 |
|
176896 |
0.03 |
| chr3_166970881_166971032 | 0.16 |
ZBBX |
zinc finger, B-box domain containing |
119820 |
0.06 |
| chr1_199582224_199582375 | 0.16 |
ENSG00000263805 |
. |
2109 |
0.48 |
| chr1_149816150_149816668 | 0.16 |
HIST2H2AA3 |
histone cluster 2, H2aa3 |
1931 |
0.11 |
| chrX_78206486_78206637 | 0.16 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
5643 |
0.34 |
| chr5_67176974_67177125 | 0.16 |
ENSG00000223149 |
. |
86224 |
0.11 |
| chr12_4384723_4384874 | 0.16 |
CCND2 |
cyclin D2 |
432 |
0.41 |
| chr22_29206752_29206905 | 0.16 |
CTA-292E10.8 |
|
4074 |
0.17 |
| chr6_26251322_26251789 | 0.16 |
HIST1H2BH |
histone cluster 1, H2bh |
324 |
0.6 |
| chr14_65734381_65734532 | 0.16 |
ENSG00000266740 |
. |
67445 |
0.11 |
| chr7_115851873_115852024 | 0.15 |
TES |
testis derived transcript (3 LIM domains) |
1345 |
0.49 |
| chr11_113644591_113644812 | 0.15 |
ZW10 |
zw10 kinetochore protein |
168 |
0.92 |
| chr1_67400477_67400653 | 0.15 |
MIER1 |
mesoderm induction early response 1, transcriptional regulator |
4639 |
0.23 |
| chr6_170862470_170862790 | 0.15 |
PSMB1 |
proteasome (prosome, macropain) subunit, beta type, 1 |
201 |
0.86 |
| chr6_159237653_159237805 | 0.15 |
EZR-AS1 |
EZR antisense RNA 1 |
1314 |
0.35 |
| chr9_276345_276496 | 0.15 |
DOCK8 |
dedicator of cytokinesis 8 |
3350 |
0.24 |
| chr1_158811509_158811660 | 0.15 |
MNDA |
myeloid cell nuclear differentiation antigen |
4107 |
0.23 |
| chr3_151963279_151963566 | 0.15 |
MBNL1 |
muscleblind-like splicing regulator 1 |
22407 |
0.21 |
| chr7_150440682_150440833 | 0.15 |
GIMAP5 |
GTPase, IMAP family member 5 |
6321 |
0.17 |
| chr4_83349699_83350038 | 0.15 |
HNRNPDL |
heterogeneous nuclear ribonucleoprotein D-like |
726 |
0.59 |
| chr1_190136920_190137071 | 0.15 |
RP11-547I7.1 |
|
97033 |
0.09 |
| chr11_34126778_34126978 | 0.15 |
NAT10 |
N-acetyltransferase 10 (GCN5-related) |
271 |
0.94 |
| chr3_16927612_16928071 | 0.15 |
PLCL2 |
phospholipase C-like 2 |
1389 |
0.54 |
| chr1_176174683_176175792 | 0.15 |
RFWD2 |
ring finger and WD repeat domain 2, E3 ubiquitin protein ligase |
280 |
0.89 |
| chrX_121293446_121293597 | 0.15 |
ENSG00000221217 |
. |
290801 |
0.01 |
| chr10_7526759_7527322 | 0.15 |
ENSG00000207453 |
. |
1731 |
0.45 |
| chr5_148725564_148725728 | 0.15 |
GRPEL2 |
GrpE-like 2, mitochondrial (E. coli) |
581 |
0.64 |
| chr19_48751391_48751542 | 0.14 |
CARD8 |
caspase recruitment domain family, member 8 |
1459 |
0.28 |
| chr16_89181351_89181649 | 0.14 |
CTD-2555A7.3 |
|
187 |
0.93 |
| chr2_119613988_119614139 | 0.14 |
EN1 |
engrailed homeobox 1 |
8809 |
0.28 |
| chr1_160676943_160677601 | 0.14 |
CD48 |
CD48 molecule |
4321 |
0.18 |
| chr6_149386973_149387124 | 0.14 |
RP11-162J8.3 |
|
33339 |
0.2 |
| chr22_32858699_32858850 | 0.14 |
BPIFC |
BPI fold containing family C |
1655 |
0.35 |
| chr10_90751494_90751719 | 0.14 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
459 |
0.66 |
| chr13_46755525_46755771 | 0.14 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
811 |
0.58 |
| chr13_75840765_75840916 | 0.14 |
ENSG00000266534 |
. |
57667 |
0.14 |
| chr3_47203422_47203573 | 0.14 |
SETD2 |
SET domain containing 2 |
1569 |
0.35 |
| chr16_25072169_25072320 | 0.14 |
ARHGAP17 |
Rho GTPase activating protein 17 |
45257 |
0.14 |
| chr1_84767342_84767579 | 0.14 |
SAMD13 |
sterile alpha motif domain containing 13 |
57 |
0.98 |
| chr15_93429972_93430811 | 0.14 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
3865 |
0.22 |
| chr4_40205321_40205527 | 0.14 |
RHOH |
ras homolog family member H |
3460 |
0.27 |
| chr7_24576141_24576292 | 0.14 |
ENSG00000206877 |
. |
735 |
0.75 |
| chr10_3514525_3515428 | 0.14 |
RP11-184A2.3 |
|
278283 |
0.01 |
| chr11_128609410_128609561 | 0.14 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
25200 |
0.18 |
| chr14_99710021_99710272 | 0.14 |
AL109767.1 |
|
19139 |
0.2 |
| chr14_58711962_58712794 | 0.14 |
PSMA3 |
proteasome (prosome, macropain) subunit, alpha type, 3 |
756 |
0.54 |
| chr4_148539335_148539606 | 0.14 |
TMEM184C |
transmembrane protein 184C |
933 |
0.44 |
| chr7_150414088_150414577 | 0.13 |
GIMAP1 |
GTPase, IMAP family member 1 |
687 |
0.65 |
| chrX_78412852_78413003 | 0.13 |
GPR174 |
G protein-coupled receptor 174 |
13542 |
0.31 |
| chr8_19613768_19613970 | 0.13 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
947 |
0.69 |
| chr17_66202564_66202990 | 0.13 |
AMZ2 |
archaelysin family metallopeptidase 2 |
40938 |
0.12 |
| chr4_146541198_146541349 | 0.13 |
MMAA |
methylmalonic aciduria (cobalamin deficiency) cblA type |
1858 |
0.37 |
| chr2_143887894_143888150 | 0.13 |
ARHGAP15 |
Rho GTPase activating protein 15 |
1139 |
0.61 |
| chr14_97925415_97925566 | 0.13 |
ENSG00000240730 |
. |
71020 |
0.14 |
| chrX_56756434_56756710 | 0.13 |
ENSG00000204272 |
. |
880 |
0.75 |
| chr1_65534522_65534674 | 0.13 |
ENSG00000199135 |
. |
10407 |
0.19 |
| chr11_66035600_66035751 | 0.13 |
RP11-867G23.2 |
|
75 |
0.76 |
| chr4_130846311_130846462 | 0.13 |
ENSG00000222861 |
. |
64731 |
0.16 |
| chr8_67974970_67975187 | 0.13 |
CSPP1 |
centrosome and spindle pole associated protein 1 |
417 |
0.53 |
| chr7_152133952_152134604 | 0.13 |
KMT2C |
lysine (K)-specific methyltransferase 2C |
1188 |
0.44 |
| chr17_10603321_10603720 | 0.13 |
ADPRM |
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
2609 |
0.18 |
| chr2_85166549_85166831 | 0.13 |
KCMF1 |
potassium channel modulatory factor 1 |
31526 |
0.15 |
| chr11_12218163_12218314 | 0.13 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
34535 |
0.18 |
| chrX_18444690_18444905 | 0.13 |
CDKL5 |
cyclin-dependent kinase-like 5 |
1094 |
0.59 |
| chr11_102724997_102725148 | 0.13 |
MMP3 |
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
10538 |
0.19 |
| chr16_71916638_71916789 | 0.13 |
RP11-498D10.3 |
|
537 |
0.48 |
| chr13_87919295_87919446 | 0.13 |
ENSG00000266052 |
. |
351625 |
0.01 |
| chr11_95881357_95881508 | 0.13 |
ENSG00000266192 |
. |
193170 |
0.03 |
| chr1_228336510_228336661 | 0.13 |
GJC2 |
gap junction protein, gamma 2, 47kDa |
968 |
0.38 |
| chr2_42979871_42980022 | 0.13 |
OXER1 |
oxoeicosanoid (OXE) receptor 1 |
11455 |
0.21 |
| chr16_71933280_71933431 | 0.13 |
IST1 |
increased sodium tolerance 1 homolog (yeast) |
3863 |
0.15 |
| chr8_29936071_29936255 | 0.13 |
TMEM66 |
transmembrane protein 66 |
3777 |
0.18 |
| chr4_25333211_25333559 | 0.13 |
ZCCHC4 |
zinc finger, CCHC domain containing 4 |
1498 |
0.44 |
| chr15_95740241_95740392 | 0.12 |
ENSG00000222076 |
. |
548717 |
0.0 |
| chr10_69609625_69609925 | 0.12 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
11853 |
0.17 |
| chr2_178187798_178187949 | 0.12 |
AC074286.1 |
|
19 |
0.96 |
| chr8_9156944_9157095 | 0.12 |
ENSG00000207415 |
. |
11931 |
0.25 |
| chr6_139458867_139459205 | 0.12 |
HECA |
headcase homolog (Drosophila) |
2787 |
0.35 |
| chr1_145507666_145508470 | 0.12 |
RP11-315I20.1 |
|
80 |
0.83 |
| chr1_31407657_31407808 | 0.12 |
ENSG00000200154 |
. |
888 |
0.51 |
| chr2_87799012_87799242 | 0.12 |
RP11-1399P15.1 |
|
21574 |
0.26 |
| chr2_201981847_201982480 | 0.12 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
401 |
0.79 |
| chr2_9315047_9315328 | 0.12 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
31707 |
0.23 |
| chr13_99992298_99992449 | 0.12 |
ENSG00000207719 |
. |
16012 |
0.19 |
| chr6_10310725_10311140 | 0.12 |
ENSG00000221583 |
. |
88663 |
0.07 |
| chr17_37356079_37356230 | 0.12 |
RPL19 |
ribosomal protein L19 |
382 |
0.77 |
| chrX_45210737_45210888 | 0.12 |
RP11-342D14.1 |
|
31109 |
0.25 |
| chr18_68289401_68289560 | 0.12 |
GTSCR1 |
Gilles de la Tourette syndrome chromosome region, candidate 1 |
28467 |
0.25 |
| chrX_48858741_48859359 | 0.12 |
GRIPAP1 |
GRIP1 associated protein 1 |
375 |
0.74 |
| chr11_58340730_58341020 | 0.12 |
LPXN |
leupaxin |
2459 |
0.23 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 | 0.3 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.1 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0035844 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.4 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) epithelial cell proliferation involved in lung morphogenesis(GO:0060502) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.0 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.0 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.0 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.0 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0032356 | single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |