Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
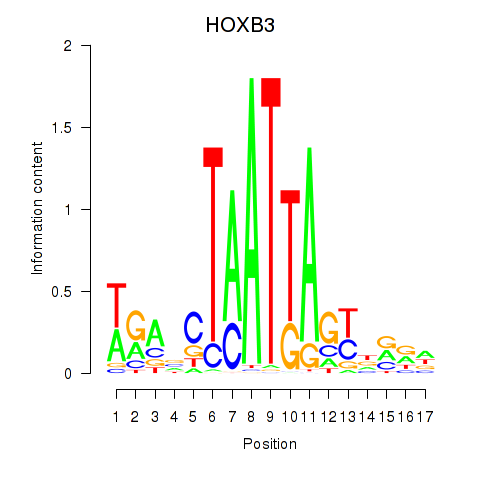
Results for HOXB3
Z-value: 1.06
Transcription factors associated with HOXB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB3
|
ENSG00000120093.7 | homeobox B3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_46643320_46643471 | HOXB3 | 7990 | 0.072051 | 0.77 | 1.6e-02 | Click! |
| chr17_46650833_46651193 | HOXB3 | 372 | 0.674742 | 0.59 | 9.8e-02 | Click! |
| chr17_46633195_46633347 | HOXB3 | 612 | 0.441121 | -0.52 | 1.5e-01 | Click! |
| chr17_46665362_46665513 | HOXB3 | 2160 | 0.112807 | -0.51 | 1.6e-01 | Click! |
| chr17_46653822_46653973 | HOXB3 | 1459 | 0.174654 | -0.47 | 2.0e-01 | Click! |
Activity of the HOXB3 motif across conditions
Conditions sorted by the z-value of the HOXB3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
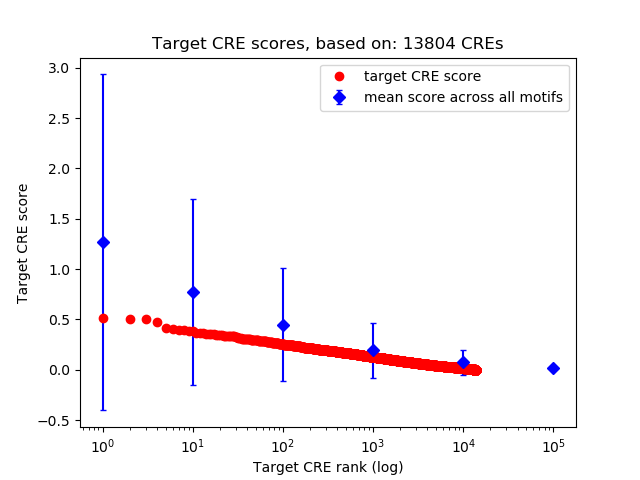
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
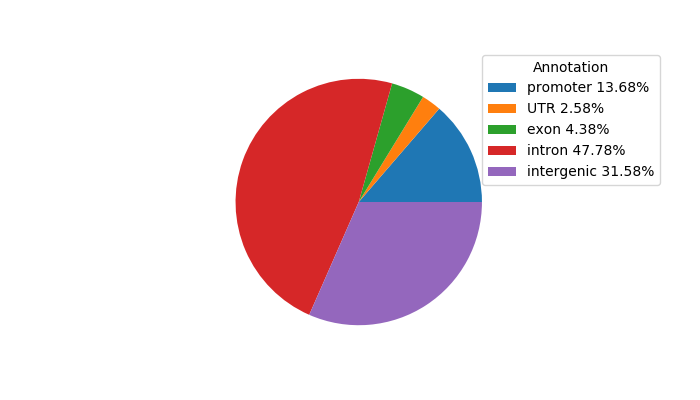
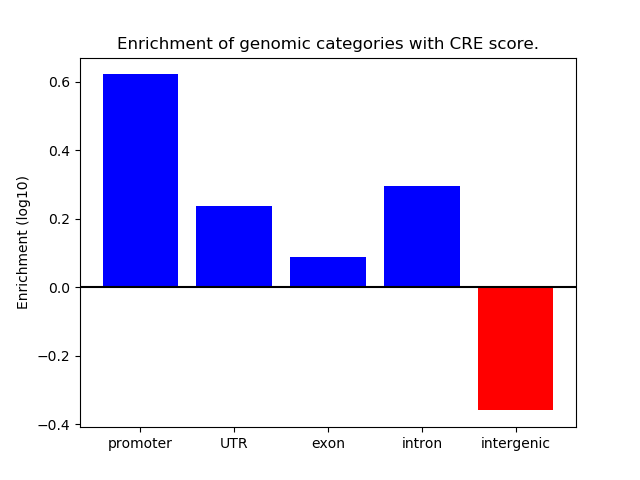
| chr2_64864648_64864996 | 0.52 |
SERTAD2 |
SERTA domain containing 2 |
16225 |
0.22 |
| chr11_128582862_128583013 | 0.51 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
17019 |
0.17 |
| chr5_147836900_147837051 | 0.50 |
CTD-2283N19.1 |
|
26605 |
0.18 |
| chr20_45946936_45948261 | 0.47 |
AL031666.2 |
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
352 |
0.82 |
| chr1_198238845_198238996 | 0.42 |
NEK7 |
NIMA-related kinase 7 |
48991 |
0.19 |
| chr3_145879077_145879748 | 0.41 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
458 |
0.86 |
| chr2_227516189_227516445 | 0.40 |
ENSG00000263363 |
. |
7192 |
0.31 |
| chr3_195273446_195273682 | 0.39 |
AC091633.3 |
|
145 |
0.95 |
| chr2_128492018_128492224 | 0.39 |
SFT2D3 |
SFT2 domain containing 3 |
33524 |
0.13 |
| chr2_153267501_153267713 | 0.39 |
FMNL2 |
formin-like 2 |
75856 |
0.12 |
| chr1_68697735_68698156 | 0.36 |
WLS |
wntless Wnt ligand secretion mediator |
18 |
0.98 |
| chr8_57517628_57517779 | 0.36 |
RP11-17A4.2 |
|
116046 |
0.06 |
| chr13_32828572_32828723 | 0.36 |
FRY |
furry homolog (Drosophila) |
10158 |
0.21 |
| chr1_151918063_151918251 | 0.36 |
THEM4 |
thioesterase superfamily member 4 |
35873 |
0.1 |
| chr7_151048885_151049126 | 0.36 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
6574 |
0.16 |
| chr20_14316163_14316462 | 0.35 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
1942 |
0.4 |
| chr14_65192350_65192501 | 0.35 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
1926 |
0.38 |
| chr10_12746098_12746480 | 0.35 |
ENSG00000221331 |
. |
21063 |
0.25 |
| chr2_63275452_63275960 | 0.35 |
AC009501.4 |
|
50 |
0.97 |
| chr9_113800702_113801367 | 0.35 |
LPAR1 |
lysophosphatidic acid receptor 1 |
53 |
0.98 |
| chr4_169756047_169756198 | 0.34 |
RP11-635L1.3 |
|
2034 |
0.31 |
| chr21_34398214_34398768 | 0.34 |
OLIG2 |
oligodendrocyte lineage transcription factor 2 |
248 |
0.93 |
| chr1_84107245_84107582 | 0.34 |
ENSG00000223231 |
. |
152147 |
0.04 |
| chr1_87799796_87800719 | 0.34 |
LMO4 |
LIM domain only 4 |
2906 |
0.4 |
| chr7_129933276_129933885 | 0.33 |
CPA4 |
carboxypeptidase A4 |
461 |
0.76 |
| chr4_40630076_40630227 | 0.33 |
RBM47 |
RNA binding motif protein 47 |
1730 |
0.47 |
| chr1_88180949_88181100 | 0.33 |
ENSG00000199318 |
. |
261968 |
0.02 |
| chr9_18976648_18977105 | 0.33 |
FAM154A |
family with sequence similarity 154, member A |
56310 |
0.1 |
| chr5_142284312_142284613 | 0.33 |
ARHGAP26 |
Rho GTPase activating protein 26 |
2425 |
0.38 |
| chr17_64187141_64188685 | 0.33 |
CEP112 |
centrosomal protein 112kDa |
60 |
0.98 |
| chr4_95367292_95367585 | 0.32 |
PDLIM5 |
PDZ and LIM domain 5 |
5599 |
0.33 |
| chr7_105912156_105912307 | 0.32 |
NAMPT |
nicotinamide phosphoribosyltransferase |
13136 |
0.21 |
| chr4_142380005_142380156 | 0.32 |
ENSG00000238695 |
. |
161048 |
0.04 |
| chr18_33554456_33554607 | 0.31 |
C18orf21 |
chromosome 18 open reading frame 21 |
1783 |
0.36 |
| chr6_16737749_16737900 | 0.31 |
RP1-151F17.1 |
|
23545 |
0.22 |
| chr7_27153769_27154326 | 0.31 |
HOXA-AS2 |
HOXA cluster antisense RNA 2 |
492 |
0.41 |
| chr12_58705170_58705321 | 0.31 |
RP11-362K2.2 |
Protein LOC100506869 |
232662 |
0.02 |
| chr8_116675050_116675578 | 0.31 |
TRPS1 |
trichorhinophalangeal syndrome I |
1409 |
0.6 |
| chr5_3597777_3597928 | 0.31 |
IRX1 |
iroquois homeobox 1 |
1684 |
0.41 |
| chr10_119303944_119304573 | 0.30 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
321 |
0.88 |
| chr13_50706829_50707141 | 0.30 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
50678 |
0.13 |
| chr14_100573958_100574109 | 0.30 |
ENSG00000265154 |
. |
1818 |
0.23 |
| chr4_139635357_139635508 | 0.30 |
ENSG00000238971 |
. |
97488 |
0.08 |
| chr19_38494188_38494741 | 0.30 |
CTC-244M17.1 |
|
39971 |
0.13 |
| chr17_30683527_30683678 | 0.30 |
ZNF207 |
zinc finger protein 207 |
5675 |
0.15 |
| chr4_160357796_160357947 | 0.30 |
ENSG00000251979 |
. |
70309 |
0.12 |
| chr20_11887627_11887778 | 0.30 |
BTBD3 |
BTB (POZ) domain containing 3 |
10863 |
0.23 |
| chr21_40357766_40357986 | 0.30 |
ENSG00000272015 |
. |
91167 |
0.09 |
| chr12_50748152_50748528 | 0.30 |
FAM186A |
family with sequence similarity 186, member A |
3672 |
0.22 |
| chr15_96878987_96879612 | 0.29 |
ENSG00000222651 |
. |
2809 |
0.21 |
| chr2_26100038_26101197 | 0.29 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
718 |
0.76 |
| chr2_207998546_207999225 | 0.29 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
24 |
0.98 |
| chr6_137544605_137544818 | 0.29 |
IFNGR1 |
interferon gamma receptor 1 |
4125 |
0.31 |
| chr18_46620789_46620940 | 0.29 |
DYM |
dymeclin |
2999 |
0.26 |
| chr18_27841796_27841947 | 0.29 |
ENSG00000251702 |
. |
722201 |
0.0 |
| chr9_17578628_17579355 | 0.29 |
SH3GL2 |
SH3-domain GRB2-like 2 |
130 |
0.98 |
| chr11_57530814_57530965 | 0.29 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
436 |
0.77 |
| chr5_35309735_35309886 | 0.29 |
PRLR |
prolactin receptor |
79016 |
0.1 |
| chr12_32635785_32635936 | 0.29 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
3046 |
0.28 |
| chr5_119820967_119821118 | 0.29 |
PRR16 |
proline rich 16 |
19716 |
0.29 |
| chr16_13401641_13401792 | 0.28 |
AC003009.1 |
|
24724 |
0.26 |
| chr12_94132438_94132603 | 0.28 |
RP11-887P2.5 |
|
921 |
0.64 |
| chr21_36254420_36254945 | 0.28 |
RUNX1 |
runt-related transcription factor 1 |
4798 |
0.34 |
| chr22_19073564_19073715 | 0.28 |
AC004471.9 |
|
35403 |
0.09 |
| chr16_17931379_17931530 | 0.28 |
XYLT1 |
xylosyltransferase I |
366716 |
0.01 |
| chr3_108988766_108988917 | 0.28 |
ENSG00000252889 |
. |
44865 |
0.15 |
| chr7_37724366_37724517 | 0.28 |
GPR141 |
G protein-coupled receptor 141 |
966 |
0.42 |
| chr12_89032903_89033122 | 0.28 |
ENSG00000252850 |
. |
57596 |
0.14 |
| chr7_32102480_32102631 | 0.28 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
7906 |
0.33 |
| chr6_50378521_50378672 | 0.28 |
TFAP2D |
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
302945 |
0.01 |
| chr3_151560621_151560772 | 0.27 |
AADAC |
arylacetamide deacetylase |
28835 |
0.17 |
| chr10_81026521_81026806 | 0.27 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
39312 |
0.18 |
| chr16_86619409_86619937 | 0.27 |
FOXL1 |
forkhead box L1 |
7558 |
0.18 |
| chr3_134050593_134051228 | 0.27 |
AMOTL2 |
angiomotin like 2 |
39844 |
0.16 |
| chr1_196577911_196578299 | 0.27 |
KCNT2 |
potassium channel, subfamily T, member 2 |
250 |
0.94 |
| chr3_33318777_33319866 | 0.27 |
FBXL2 |
F-box and leucine-rich repeat protein 2 |
353 |
0.91 |
| chr11_74292413_74292564 | 0.27 |
POLD3 |
polymerase (DNA-directed), delta 3, accessory subunit |
11087 |
0.2 |
| chr3_69434805_69435778 | 0.27 |
FRMD4B |
FERM domain containing 4B |
67 |
0.99 |
| chr10_4170534_4170685 | 0.27 |
KLF6 |
Kruppel-like factor 6 |
343136 |
0.01 |
| chr2_58266503_58266768 | 0.27 |
VRK2 |
vaccinia related kinase 2 |
7094 |
0.27 |
| chr1_95330057_95330663 | 0.27 |
SLC44A3 |
solute carrier family 44, member 3 |
2527 |
0.28 |
| chr15_43985556_43986365 | 0.27 |
CKMT1A |
creatine kinase, mitochondrial 1A |
109 |
0.93 |
| chr1_52605764_52605957 | 0.27 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
2186 |
0.28 |
| chr18_45480065_45480352 | 0.27 |
SMAD2 |
SMAD family member 2 |
22693 |
0.23 |
| chr4_89714293_89714444 | 0.27 |
FAM13A |
family with sequence similarity 13, member A |
29984 |
0.19 |
| chr12_46949518_46949939 | 0.26 |
SLC38A2 |
solute carrier family 38, member 2 |
183078 |
0.03 |
| chr1_225842736_225843127 | 0.26 |
ENAH |
enabled homolog (Drosophila) |
2087 |
0.34 |
| chr2_191624663_191624826 | 0.26 |
AC006460.2 |
|
51302 |
0.14 |
| chr19_58488964_58489115 | 0.26 |
C19orf18 |
chromosome 19 open reading frame 18 |
3137 |
0.15 |
| chr10_75668708_75669249 | 0.26 |
PLAU |
plasminogen activator, urokinase |
43 |
0.97 |
| chr13_76026824_76027007 | 0.26 |
TBC1D4 |
TBC1 domain family, member 4 |
29335 |
0.19 |
| chr3_168240363_168240514 | 0.26 |
ENSG00000207717 |
. |
29204 |
0.22 |
| chr2_195207760_195207911 | 0.26 |
ENSG00000264560 |
. |
215163 |
0.02 |
| chr2_197103959_197104132 | 0.26 |
AC020571.3 |
|
20703 |
0.17 |
| chr5_159524987_159525138 | 0.26 |
PWWP2A |
PWWP domain containing 2A |
21367 |
0.17 |
| chrX_41379072_41379223 | 0.26 |
CASK-AS1 |
CASK antisense RNA 1 |
142 |
0.96 |
| chr9_20490881_20491032 | 0.26 |
ENSG00000264941 |
. |
11384 |
0.23 |
| chr12_46759532_46760006 | 0.26 |
SLC38A2 |
solute carrier family 38, member 2 |
679 |
0.75 |
| chr6_58148368_58148782 | 0.26 |
ENSG00000212017 |
. |
893645 |
0.0 |
| chr10_33798417_33798757 | 0.25 |
NRP1 |
neuropilin 1 |
173397 |
0.03 |
| chr3_65949778_65949974 | 0.25 |
MAGI1-IT1 |
MAGI1 intronic transcript 1 (non-protein coding) |
9643 |
0.19 |
| chr12_65700600_65700821 | 0.25 |
MSRB3 |
methionine sulfoxide reductase B3 |
19945 |
0.21 |
| chr19_30436184_30436558 | 0.25 |
URI1 |
URI1, prefoldin-like chaperone |
2946 |
0.39 |
| chr1_209975427_209975724 | 0.25 |
IRF6 |
interferon regulatory factor 6 |
64 |
0.96 |
| chr12_18414636_18414918 | 0.25 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
303 |
0.95 |
| chr12_115135696_115136278 | 0.25 |
TBX3 |
T-box 3 |
14018 |
0.21 |
| chr22_38723559_38723988 | 0.25 |
CSNK1E |
casein kinase 1, epsilon |
9684 |
0.15 |
| chr20_10502111_10502382 | 0.25 |
SLX4IP |
SLX4 interacting protein |
86295 |
0.09 |
| chr3_108553094_108553245 | 0.25 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
11550 |
0.26 |
| chr12_106699376_106699527 | 0.25 |
CKAP4 |
cytoskeleton-associated protein 4 |
1394 |
0.34 |
| chrX_11456470_11456766 | 0.25 |
ARHGAP6 |
Rho GTPase activating protein 6 |
10725 |
0.3 |
| chr10_45389082_45389233 | 0.25 |
TMEM72 |
transmembrane protein 72 |
17491 |
0.16 |
| chr11_33466056_33466207 | 0.25 |
ENSG00000223134 |
. |
90120 |
0.08 |
| chr3_9006349_9006500 | 0.25 |
RAD18 |
RAD18 homolog (S. cerevisiae) |
967 |
0.6 |
| chr2_103506378_103506656 | 0.25 |
TMEM182 |
transmembrane protein 182 |
128045 |
0.06 |
| chr18_77385131_77385282 | 0.25 |
CTDP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
54595 |
0.14 |
| chr16_3911841_3912078 | 0.25 |
CREBBP |
CREB binding protein |
18162 |
0.21 |
| chr15_79172438_79172758 | 0.25 |
MORF4L1 |
mortality factor 4 like 1 |
2016 |
0.31 |
| chr12_51640112_51640647 | 0.25 |
DAZAP2 |
DAZ associated protein 2 |
7300 |
0.14 |
| chr4_47544899_47545050 | 0.25 |
AC092597.3 |
|
17302 |
0.23 |
| chr1_3736530_3736681 | 0.25 |
CEP104 |
centrosomal protein 104kDa |
9265 |
0.12 |
| chr8_79260999_79261150 | 0.25 |
ENSG00000252935 |
. |
49783 |
0.19 |
| chr7_83932213_83932364 | 0.25 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
107527 |
0.08 |
| chr2_176709130_176709424 | 0.25 |
AC016751.3 |
|
52787 |
0.15 |
| chr2_158039761_158039912 | 0.25 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
74274 |
0.11 |
| chr19_53716573_53716772 | 0.24 |
ZNF665 |
zinc finger protein 665 |
20045 |
0.1 |
| chrX_64908310_64908461 | 0.24 |
MSN |
moesin |
20848 |
0.28 |
| chr6_139292312_139292463 | 0.24 |
REPS1 |
RALBP1 associated Eps domain containing 1 |
16390 |
0.23 |
| chr10_114714851_114715858 | 0.24 |
RP11-57H14.2 |
|
3720 |
0.26 |
| chr4_177931414_177931624 | 0.24 |
ENSG00000222859 |
. |
171077 |
0.03 |
| chr7_123390243_123390394 | 0.24 |
RP11-390E23.6 |
|
1196 |
0.34 |
| chr5_14153831_14154379 | 0.24 |
TRIO |
trio Rho guanine nucleotide exchange factor |
10276 |
0.32 |
| chr8_41165312_41165463 | 0.24 |
SFRP1 |
secreted frizzled-related protein 1 |
878 |
0.57 |
| chr6_133066944_133067095 | 0.24 |
RP1-55C23.7 |
|
6795 |
0.13 |
| chr5_77886889_77887187 | 0.24 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
42064 |
0.21 |
| chr15_43885730_43886468 | 0.24 |
CKMT1B |
creatine kinase, mitochondrial 1B |
42 |
0.95 |
| chr2_158113918_158114875 | 0.24 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
286 |
0.93 |
| chr10_33422709_33422860 | 0.24 |
ENSG00000263576 |
. |
35220 |
0.17 |
| chr19_39143944_39144135 | 0.24 |
ACTN4 |
actinin, alpha 4 |
5661 |
0.13 |
| chr13_40858172_40858323 | 0.24 |
ENSG00000207458 |
. |
57283 |
0.16 |
| chr1_51069078_51069376 | 0.24 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
8943 |
0.23 |
| chr8_49612409_49612560 | 0.24 |
EFCAB1 |
EF-hand calcium binding domain 1 |
29864 |
0.25 |
| chr10_31273151_31273302 | 0.24 |
ZNF438 |
zinc finger protein 438 |
129 |
0.98 |
| chr6_157381110_157381404 | 0.24 |
RP1-137K2.2 |
|
59583 |
0.15 |
| chr2_228735999_228736581 | 0.23 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
33 |
0.98 |
| chr11_22724799_22724998 | 0.23 |
ENSG00000222427 |
. |
15001 |
0.2 |
| chr20_56749874_56750278 | 0.23 |
C20orf85 |
chromosome 20 open reading frame 85 |
24116 |
0.19 |
| chr15_82414957_82415108 | 0.23 |
RP11-597K23.2 |
Uncharacterized protein |
34000 |
0.16 |
| chr13_37397601_37397752 | 0.23 |
RFXAP |
regulatory factor X-associated protein |
4315 |
0.24 |
| chr20_52370901_52371111 | 0.23 |
ENSG00000238468 |
. |
85709 |
0.09 |
| chr1_202438325_202438476 | 0.23 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
6529 |
0.23 |
| chr13_100860702_100860853 | 0.23 |
PCCA |
propionyl CoA carboxylase, alpha polypeptide |
84701 |
0.1 |
| chr2_188360675_188360826 | 0.23 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
17618 |
0.23 |
| chr4_156590981_156591132 | 0.23 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
2241 |
0.4 |
| chr4_114705923_114706074 | 0.23 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
22915 |
0.27 |
| chr17_33391542_33391948 | 0.23 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
874 |
0.45 |
| chr3_162506552_162506703 | 0.23 |
ENSG00000238398 |
. |
390136 |
0.01 |
| chr12_78336292_78336638 | 0.23 |
NAV3 |
neuron navigator 3 |
23591 |
0.28 |
| chr20_35924690_35924841 | 0.23 |
MANBAL |
mannosidase, beta A, lysosomal-like |
534 |
0.79 |
| chr18_55891591_55891981 | 0.23 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
2983 |
0.31 |
| chr8_57086542_57086822 | 0.23 |
PLAG1 |
pleiomorphic adenoma gene 1 |
37156 |
0.12 |
| chr2_147107195_147107346 | 0.23 |
ENSG00000251905 |
. |
204546 |
0.03 |
| chr9_79483882_79484033 | 0.23 |
PRUNE2 |
prune homolog 2 (Drosophila) |
37044 |
0.21 |
| chr1_200987607_200987758 | 0.23 |
KIF21B |
kinesin family member 21B |
4854 |
0.21 |
| chr1_181069341_181069492 | 0.23 |
IER5 |
immediate early response 5 |
11778 |
0.23 |
| chr5_123252700_123253101 | 0.23 |
CSNK1G3 |
casein kinase 1, gamma 3 |
328781 |
0.01 |
| chr6_83094369_83094520 | 0.22 |
TPBG |
trophoblast glycoprotein |
20483 |
0.28 |
| chr6_1614659_1614953 | 0.22 |
FOXC1 |
forkhead box C1 |
4125 |
0.35 |
| chr2_195590152_195590303 | 0.22 |
ENSG00000252517 |
. |
61329 |
0.16 |
| chr7_92254665_92254816 | 0.22 |
FAM133B |
family with sequence similarity 133, member B |
35032 |
0.17 |
| chr21_19152947_19153098 | 0.22 |
AL109761.5 |
|
12783 |
0.23 |
| chr4_15963408_15963559 | 0.22 |
FGFBP2 |
fibroblast growth factor binding protein 2 |
1376 |
0.42 |
| chr12_96094020_96094171 | 0.22 |
ENSG00000212448 |
. |
82630 |
0.07 |
| chr12_27901688_27901839 | 0.22 |
ENSG00000252585 |
. |
4044 |
0.14 |
| chr5_73839186_73839374 | 0.22 |
HEXB |
hexosaminidase B (beta polypeptide) |
96568 |
0.07 |
| chr15_30112674_30113382 | 0.22 |
TJP1 |
tight junction protein 1 |
723 |
0.64 |
| chr4_48240161_48240435 | 0.22 |
TEC |
tec protein tyrosine kinase |
31583 |
0.18 |
| chr7_93003610_93003782 | 0.22 |
CCDC132 |
coiled-coil domain containing 132 |
24486 |
0.23 |
| chr7_115670709_115671033 | 0.22 |
TFEC |
transcription factor EC |
4 |
0.99 |
| chr4_459906_460057 | 0.22 |
ENSG00000252150 |
. |
1444 |
0.36 |
| chr5_17223836_17223987 | 0.22 |
BASP1 |
brain abundant, membrane attached signal protein 1 |
6242 |
0.19 |
| chr4_141984739_141984890 | 0.22 |
RNF150 |
ring finger protein 150 |
9934 |
0.3 |
| chr2_43822335_43823890 | 0.22 |
THADA |
thyroid adenoma associated |
1 |
0.98 |
| chr1_189407105_189407256 | 0.22 |
ENSG00000252553 |
. |
228206 |
0.02 |
| chr17_39742070_39742221 | 0.22 |
KRT14 |
keratin 14 |
1028 |
0.36 |
| chr5_73536836_73537130 | 0.22 |
ENSG00000222551 |
. |
30607 |
0.23 |
| chrX_39548256_39548819 | 0.22 |
ENSG00000263730 |
. |
28067 |
0.24 |
| chr9_96819103_96819334 | 0.22 |
PTPDC1 |
protein tyrosine phosphatase domain containing 1 |
26142 |
0.21 |
| chr15_71606160_71606311 | 0.22 |
RP11-592N21.2 |
|
28428 |
0.23 |
| chr7_38546645_38546796 | 0.22 |
ENSG00000241756 |
. |
24219 |
0.23 |
| chr11_128453017_128453335 | 0.22 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
4277 |
0.28 |
| chr15_34875013_34876331 | 0.22 |
GOLGA8B |
golgin A8 family, member B |
99 |
0.96 |
| chr6_7727536_7728016 | 0.22 |
BMP6 |
bone morphogenetic protein 6 |
746 |
0.78 |
| chr13_52108240_52108391 | 0.22 |
INTS6-AS1 |
INTS6 antisense RNA 1 |
7379 |
0.15 |
| chr10_79470838_79471281 | 0.22 |
ENSG00000199664 |
. |
65754 |
0.1 |
| chr22_46436012_46436163 | 0.22 |
RP6-109B7.5 |
|
12886 |
0.1 |
| chrX_95690881_95691032 | 0.22 |
ENSG00000223260 |
. |
25175 |
0.28 |
| chr4_88817282_88817552 | 0.22 |
MEPE |
matrix extracellular phosphoglycoprotein |
63278 |
0.1 |
| chr3_71591333_71591803 | 0.21 |
ENSG00000221264 |
. |
328 |
0.83 |
| chr9_89952337_89952906 | 0.21 |
ENSG00000212421 |
. |
77256 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.1 | 0.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.3 | GO:0060013 | righting reflex(GO:0060013) |
| 0.1 | 0.1 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.1 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.2 | GO:0032344 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.1 | 0.2 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.3 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.2 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.1 | 0.2 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.3 | GO:0048875 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 0.2 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.2 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0052169 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.3 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.1 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.4 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.0 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0006991 | response to sterol depletion(GO:0006991) |
| 0.0 | 0.1 | GO:0010665 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0003160 | endocardium development(GO:0003157) endocardium morphogenesis(GO:0003160) endocardium formation(GO:0060214) |
| 0.0 | 0.3 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0002604 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0001946 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 | 0.0 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.0 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) |
| 0.0 | 0.1 | GO:0072666 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0045425 | interleukin-3 production(GO:0032632) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.2 | GO:0001738 | morphogenesis of a polarized epithelium(GO:0001738) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.0 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.0 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.2 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.0 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0019359 | nicotinamide nucleotide biosynthetic process(GO:0019359) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.0 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.2 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.0 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.3 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.3 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.1 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.2 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) arachidonic acid binding(GO:0050544) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.0 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |