Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
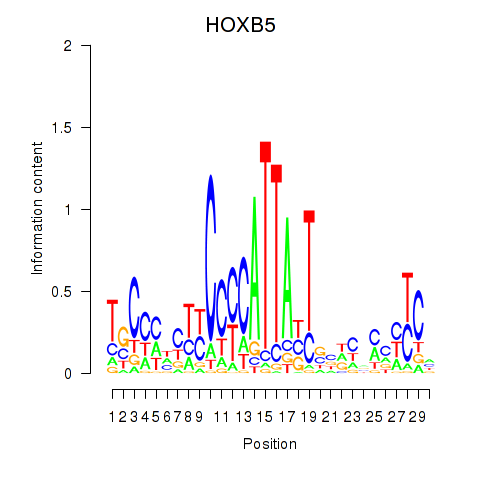
Results for HOXB5
Z-value: 0.76
Transcription factors associated with HOXB5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB5
|
ENSG00000120075.5 | homeobox B5 |
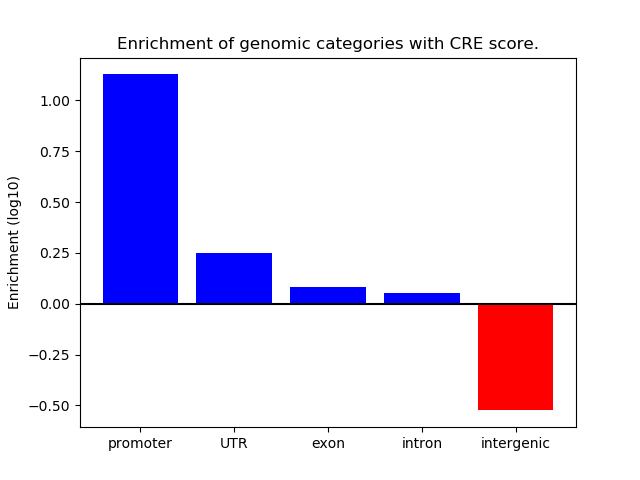
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_46672091_46672242 | HOXB5 | 843 | 0.283585 | 0.21 | 5.9e-01 | Click! |
| chr17_46671860_46672011 | HOXB5 | 612 | 0.419693 | 0.18 | 6.4e-01 | Click! |
| chr17_46671414_46671766 | HOXB5 | 267 | 0.750635 | -0.05 | 9.1e-01 | Click! |
| chr17_46670468_46671125 | HOXB5 | 527 | 0.463731 | -0.02 | 9.7e-01 | Click! |
Activity of the HOXB5 motif across conditions
Conditions sorted by the z-value of the HOXB5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
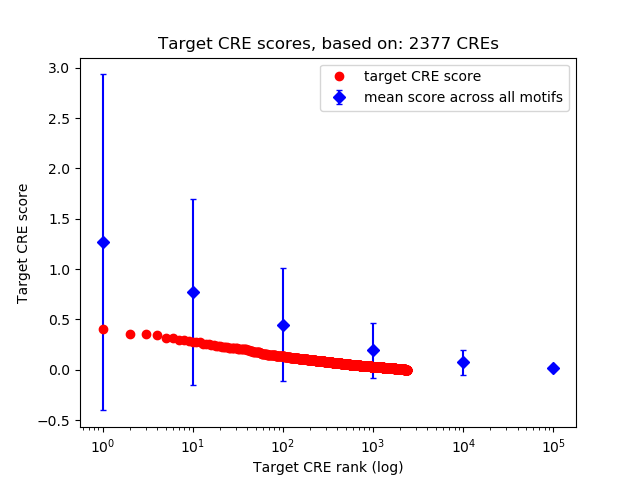
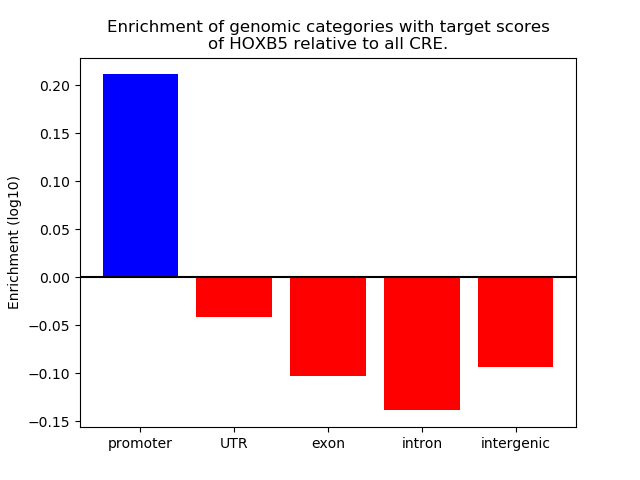
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_150536014_150536165 | 0.41 |
ANXA6 |
annexin A6 |
1219 |
0.5 |
| chr19_17958305_17958511 | 0.36 |
JAK3 |
Janus kinase 3 |
418 |
0.74 |
| chr15_93276931_93277107 | 0.35 |
CTD-2313J17.5 |
|
47703 |
0.14 |
| chr10_22541853_22542149 | 0.35 |
EBLN1 |
endogenous Bornavirus-like nucleoprotein 1 |
43051 |
0.13 |
| chr1_225655628_225655947 | 0.32 |
RP11-496N12.6 |
|
2742 |
0.3 |
| chr1_88150357_88150552 | 0.31 |
ENSG00000199318 |
. |
231398 |
0.02 |
| chr5_14441548_14441780 | 0.30 |
TRIO |
trio Rho guanine nucleotide exchange factor |
46893 |
0.19 |
| chrX_19817156_19817441 | 0.30 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
571 |
0.85 |
| chr15_89349083_89349246 | 0.29 |
ACAN |
aggrecan |
2174 |
0.38 |
| chr7_39170524_39170675 | 0.28 |
POU6F2 |
POU class 6 homeobox 2 |
45138 |
0.18 |
| chr1_228075022_228075233 | 0.28 |
ENSG00000264483 |
. |
54257 |
0.1 |
| chr19_14143598_14143749 | 0.28 |
IL27RA |
interleukin 27 receptor, alpha |
1113 |
0.26 |
| chrX_57618068_57618312 | 0.26 |
ZXDB |
zinc finger, X-linked, duplicated B |
79 |
0.99 |
| chr20_45333994_45334145 | 0.26 |
SLC2A10 |
solute carrier family 2 (facilitated glucose transporter), member 10 |
4057 |
0.22 |
| chr10_61667583_61667734 | 0.25 |
CCDC6 |
coiled-coil domain containing 6 |
1244 |
0.57 |
| chr11_17671771_17671922 | 0.24 |
RP11-358H18.2 |
|
46641 |
0.13 |
| chr12_12602626_12602777 | 0.24 |
DUSP16 |
dual specificity phosphatase 16 |
71358 |
0.09 |
| chr3_115503593_115504040 | 0.24 |
ENSG00000243359 |
. |
52899 |
0.17 |
| chr11_128370521_128370777 | 0.24 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
4640 |
0.28 |
| chr15_65903705_65903940 | 0.24 |
SLC24A1 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
79 |
0.66 |
| chr5_121836809_121836973 | 0.23 |
CTC-210G5.1 |
|
22109 |
0.24 |
| chr11_113929660_113929818 | 0.23 |
ZBTB16 |
zinc finger and BTB domain containing 16 |
576 |
0.79 |
| chr12_88812261_88812515 | 0.23 |
ENSG00000199245 |
. |
11831 |
0.3 |
| chr13_58205325_58205914 | 0.22 |
PCDH17 |
protocadherin 17 |
1036 |
0.71 |
| chr20_1637966_1638117 | 0.22 |
SIRPG |
signal-regulatory protein gamma |
319 |
0.86 |
| chr1_46632976_46633201 | 0.22 |
TSPAN1 |
tetraspanin 1 |
7671 |
0.13 |
| chr1_150124988_150125267 | 0.22 |
PLEKHO1 |
pleckstrin homology domain containing, family O member 1 |
2957 |
0.21 |
| chr5_118309425_118309661 | 0.21 |
DTWD2 |
DTW domain containing 2 |
14372 |
0.2 |
| chr1_59522819_59522970 | 0.21 |
FGGY |
FGGY carbohydrate kinase domain containing |
239416 |
0.02 |
| chr12_88811798_88812120 | 0.21 |
ENSG00000199245 |
. |
12260 |
0.3 |
| chr9_36191581_36192111 | 0.21 |
CLTA |
clathrin, light chain A |
886 |
0.58 |
| chr1_157963513_157964097 | 0.21 |
KIRREL |
kin of IRRE like (Drosophila) |
370 |
0.89 |
| chr1_226863033_226863244 | 0.21 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
370 |
0.88 |
| chrX_1653880_1654070 | 0.21 |
P2RY8 |
purinergic receptor P2Y, G-protein coupled, 8 |
2025 |
0.36 |
| chrY_1603880_1604065 | 0.21 |
NA |
NA |
> 106 |
NA |
| chr2_74758029_74758308 | 0.21 |
HTRA2 |
HtrA serine peptidase 2 |
995 |
0.23 |
| chr12_14518062_14518320 | 0.21 |
ATF7IP |
activating transcription factor 7 interacting protein |
425 |
0.88 |
| chr1_243416937_243417168 | 0.20 |
CEP170 |
centrosomal protein 170kDa |
430 |
0.81 |
| chr7_102036309_102036664 | 0.20 |
PRKRIP1 |
PRKR interacting protein 1 (IL11 inducible) |
324 |
0.8 |
| chrX_39953289_39953491 | 0.20 |
BCOR |
BCL6 corepressor |
3266 |
0.39 |
| chr2_198160747_198160898 | 0.19 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
6421 |
0.18 |
| chr14_51027599_51027854 | 0.19 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
118 |
0.91 |
| chrX_20327856_20328007 | 0.19 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
42408 |
0.18 |
| chr10_97415335_97415893 | 0.19 |
ALDH18A1 |
aldehyde dehydrogenase 18 family, member A1 |
849 |
0.66 |
| chr3_128493983_128494134 | 0.19 |
ENSG00000244232 |
. |
9932 |
0.18 |
| chr4_68411509_68411863 | 0.19 |
CENPC |
centromere protein C |
362 |
0.89 |
| chr1_26644827_26645344 | 0.18 |
UBXN11 |
UBX domain protein 11 |
231 |
0.76 |
| chrX_152600602_152600753 | 0.18 |
ZNF275 |
zinc finger protein 275 |
1064 |
0.49 |
| chr9_45781764_45781915 | 0.18 |
ENSG00000238930 |
. |
26489 |
0.19 |
| chr6_110500956_110501177 | 0.18 |
WASF1 |
WAS protein family, member 1 |
60 |
0.78 |
| chr5_175621026_175621202 | 0.18 |
SIMC1 |
SUMO-interacting motifs containing 1 |
44251 |
0.14 |
| chrX_55545389_55545540 | 0.18 |
USP51 |
ubiquitin specific peptidase 51 |
29829 |
0.17 |
| chr12_7014349_7014709 | 0.17 |
LRRC23 |
leucine rich repeat containing 23 |
269 |
0.76 |
| chr5_98126794_98126945 | 0.17 |
RGMB |
repulsive guidance molecule family member b |
17530 |
0.21 |
| chr18_60825225_60825488 | 0.17 |
RP11-299P2.1 |
|
6803 |
0.25 |
| chr7_50344203_50344943 | 0.17 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
195 |
0.97 |
| chr5_139031248_139031399 | 0.17 |
CXXC5 |
CXXC finger protein 5 |
749 |
0.7 |
| chr4_175056009_175056160 | 0.16 |
FBXO8 |
F-box protein 8 |
148730 |
0.04 |
| chr5_53765584_53765762 | 0.16 |
HSPB3 |
heat shock 27kDa protein 3 |
14228 |
0.25 |
| chr1_200639269_200639714 | 0.16 |
DDX59 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 |
394 |
0.89 |
| chr8_122583317_122583658 | 0.16 |
ENSG00000263525 |
. |
28252 |
0.22 |
| chr5_38445912_38446355 | 0.16 |
EGFLAM |
EGF-like, fibronectin type III and laminin G domains |
435 |
0.81 |
| chr11_19798727_19799172 | 0.16 |
NAV2 |
neuron navigator 2 |
64 |
0.98 |
| chr9_67737610_67737761 | 0.16 |
ENSG00000199432 |
. |
26837 |
0.15 |
| chr1_212112149_212112479 | 0.16 |
ENSG00000212187 |
. |
86589 |
0.07 |
| chr9_4661797_4662588 | 0.15 |
PPAPDC2 |
phosphatidic acid phosphatase type 2 domain containing 2 |
106 |
0.96 |
| chr9_46168095_46168246 | 0.15 |
ENSG00000238413 |
. |
26495 |
0.23 |
| chr4_149363777_149364242 | 0.15 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
337 |
0.94 |
| chr22_30641304_30641455 | 0.15 |
RP1-102K2.8 |
|
605 |
0.52 |
| chr1_153965101_153965403 | 0.15 |
RPS27 |
ribosomal protein S27 |
2014 |
0.15 |
| chr9_97021933_97022185 | 0.15 |
ZNF169 |
zinc finger protein 169 |
449 |
0.8 |
| chr3_164914749_164914991 | 0.15 |
SLITRK3 |
SLIT and NTRK-like family, member 3 |
27 |
0.99 |
| chr22_30822061_30822534 | 0.15 |
MTFP1 |
mitochondrial fission process 1 |
492 |
0.57 |
| chr8_82193202_82194095 | 0.15 |
RP11-363E6.3 |
|
33 |
0.92 |
| chr9_45047006_45047157 | 0.15 |
ENSG00000238749 |
. |
27178 |
0.23 |
| chr4_3304601_3305007 | 0.15 |
RGS12 |
regulator of G-protein signaling 12 |
5958 |
0.26 |
| chr19_24216997_24217256 | 0.14 |
ZNF254 |
zinc finger protein 254 |
850 |
0.66 |
| chr3_129205530_129205703 | 0.14 |
IFT122 |
intraflagellar transport 122 homolog (Chlamydomonas) |
1418 |
0.36 |
| chr16_56709683_56709834 | 0.14 |
MT1H |
metallothionein 1H |
6006 |
0.08 |
| chr1_16276329_16276480 | 0.14 |
ZBTB17 |
zinc finger and BTB domain containing 17 |
5481 |
0.16 |
| chr13_48795647_48795882 | 0.14 |
ITM2B |
integral membrane protein 2B |
11530 |
0.26 |
| chr9_46336000_46336151 | 0.14 |
ENSG00000239090 |
. |
26530 |
0.17 |
| chr10_29607372_29607594 | 0.14 |
LYZL1 |
lysozyme-like 1 |
29493 |
0.23 |
| chr12_6771613_6772265 | 0.14 |
ING4 |
inhibitor of growth family, member 4 |
310 |
0.57 |
| chr14_64009252_64009486 | 0.14 |
PPP2R5E |
protein phosphatase 2, regulatory subunit B', epsilon isoform |
647 |
0.51 |
| chr7_100181255_100181941 | 0.14 |
FBXO24 |
F-box protein 24 |
7 |
0.93 |
| chr14_22314825_22314976 | 0.14 |
ENSG00000222776 |
. |
66115 |
0.13 |
| chr6_95345075_95345226 | 0.14 |
MANEA-AS1 |
MANEA antisense RNA 1 (head to head) |
680176 |
0.0 |
| chr1_228652543_228652819 | 0.14 |
ENSG00000266174 |
. |
2906 |
0.12 |
| chr11_117102027_117102760 | 0.14 |
PCSK7 |
proprotein convertase subtilisin/kexin type 7 |
191 |
0.87 |
| chr1_57039327_57039478 | 0.14 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
5839 |
0.3 |
| chr2_8747464_8747615 | 0.14 |
AC011747.7 |
|
68357 |
0.12 |
| chr18_47339976_47340201 | 0.14 |
ACAA2 |
acetyl-CoA acyltransferase 2 |
219 |
0.45 |
| chr2_207938936_207939147 | 0.14 |
ENSG00000253008 |
. |
35756 |
0.16 |
| chr2_65219193_65219344 | 0.14 |
ENSG00000252892 |
. |
2612 |
0.22 |
| chr16_14014863_14015110 | 0.13 |
ERCC4 |
excision repair cross-complementing rodent repair deficiency, complementation group 4 |
972 |
0.64 |
| chr19_13262272_13262447 | 0.13 |
IER2 |
immediate early response 2 |
430 |
0.63 |
| chr6_3119416_3119591 | 0.13 |
BPHL |
biphenyl hydrolase-like (serine hydrolase) |
577 |
0.68 |
| chr1_145472532_145473065 | 0.13 |
ANKRD34A |
ankyrin repeat domain 34A |
2290 |
0.15 |
| chr2_98206021_98206285 | 0.13 |
ANKRD36B |
ankyrin repeat domain 36B |
31 |
0.98 |
| chr2_71175523_71175730 | 0.13 |
AC007040.7 |
|
56 |
0.94 |
| chr15_101667565_101667737 | 0.13 |
RP11-505E24.2 |
|
41380 |
0.16 |
| chr11_47430197_47430394 | 0.13 |
SLC39A13 |
solute carrier family 39 (zinc transporter), member 13 |
113 |
0.75 |
| chr6_4134531_4135049 | 0.13 |
RP3-400B16.4 |
|
867 |
0.4 |
| chr11_114271389_114271817 | 0.13 |
RBM7 |
RNA binding motif protein 7 |
207 |
0.74 |
| chr1_95389567_95389941 | 0.13 |
CNN3 |
calponin 3, acidic |
1583 |
0.36 |
| chr20_19991582_19991882 | 0.13 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
6028 |
0.22 |
| chr7_55433758_55433937 | 0.13 |
LANCL2 |
LanC lantibiotic synthetase component C-like 2 (bacterial) |
706 |
0.8 |
| chr19_46498683_46499250 | 0.13 |
CCDC61 |
coiled-coil domain containing 61 |
220 |
0.9 |
| chr19_52592991_52593142 | 0.13 |
ZNF841 |
zinc finger protein 841 |
4292 |
0.14 |
| chrX_52964079_52964300 | 0.13 |
FAM156A |
family with sequence similarity 156, member A |
21340 |
0.17 |
| chr16_74640360_74640757 | 0.13 |
GLG1 |
golgi glycoprotein 1 |
434 |
0.86 |
| chr8_134511768_134511919 | 0.13 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
217 |
0.97 |
| chr5_114958288_114958558 | 0.13 |
TMED7-TICAM2 |
TMED7-TICAM2 readthrough |
3288 |
0.18 |
| chr9_101848573_101848724 | 0.13 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
17672 |
0.22 |
| chr1_85086822_85086974 | 0.13 |
C1orf180 |
chromosome 1 open reading frame 180 |
10533 |
0.21 |
| chr5_174906870_174907021 | 0.13 |
SFXN1 |
sideroflexin 1 |
1350 |
0.48 |
| chr2_10445528_10445697 | 0.12 |
HPCAL1 |
hippocalcin-like 1 |
1786 |
0.33 |
| chr3_149285300_149285451 | 0.12 |
WWTR1 |
WW domain containing transcription regulator 1 |
8664 |
0.21 |
| chr11_96073941_96074227 | 0.12 |
ENSG00000266192 |
. |
518 |
0.76 |
| chr7_29899550_29899701 | 0.12 |
WIPF3 |
WAS/WASL interacting protein family, member 3 |
25284 |
0.19 |
| chrX_106872615_106872887 | 0.12 |
PRPS1 |
phosphoribosyl pyrophosphate synthetase 1 |
958 |
0.65 |
| chr7_100143879_100144474 | 0.12 |
AGFG2 |
ArfGAP with FG repeats 2 |
7294 |
0.09 |
| chr3_45430392_45430595 | 0.12 |
LARS2 |
leucyl-tRNA synthetase 2, mitochondrial |
382 |
0.91 |
| chrX_70324263_70324497 | 0.12 |
CXorf65 |
chromosome X open reading frame 65 |
2075 |
0.18 |
| chr21_40983827_40983978 | 0.12 |
C21orf88 |
chromosome 21 open reading frame 88 |
466 |
0.83 |
| chr12_98990277_98990538 | 0.12 |
SLC25A3 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 |
2904 |
0.18 |
| chr2_227476608_227476759 | 0.12 |
ENSG00000263363 |
. |
46826 |
0.19 |
| chr8_101271300_101271587 | 0.12 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
44015 |
0.13 |
| chr5_157171107_157171579 | 0.12 |
LSM11 |
LSM11, U7 small nuclear RNA associated |
640 |
0.64 |
| chr14_24641513_24641917 | 0.12 |
REC8 |
REC8 meiotic recombination protein |
105 |
0.89 |
| chr2_192116601_192116752 | 0.12 |
MYO1B |
myosin IB |
5667 |
0.29 |
| chr18_54046096_54046251 | 0.12 |
TXNL1 |
thioredoxin-like 1 |
235549 |
0.02 |
| chr6_35267664_35267815 | 0.12 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
2110 |
0.3 |
| chr2_208757346_208757497 | 0.12 |
AC083900.1 |
|
22782 |
0.19 |
| chr20_25175950_25176201 | 0.12 |
ENTPD6 |
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
254 |
0.92 |
| chr1_156184119_156184270 | 0.12 |
PMF1-BGLAP |
PMF1-BGLAP readthrough |
1394 |
0.21 |
| chr11_2966476_2966712 | 0.12 |
PHLDA2 |
pleckstrin homology-like domain, family A, member 2 |
15909 |
0.11 |
| chr12_2943957_2944189 | 0.12 |
NRIP2 |
nuclear receptor interacting protein 2 |
117 |
0.93 |
| chr20_31069881_31070147 | 0.12 |
C20orf112 |
chromosome 20 open reading frame 112 |
1260 |
0.46 |
| chr1_53662768_53663349 | 0.12 |
CPT2 |
carnitine palmitoyltransferase 2 |
957 |
0.4 |
| chr6_34855495_34855727 | 0.11 |
TAF11 |
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa |
208 |
0.91 |
| chr3_115342492_115342792 | 0.11 |
GAP43 |
growth associated protein 43 |
285 |
0.94 |
| chr12_68041813_68042274 | 0.11 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
75 |
0.98 |
| chr2_103034093_103034244 | 0.11 |
IL18RAP |
interleukin 18 receptor accessory protein |
981 |
0.47 |
| chr12_123714056_123714279 | 0.11 |
MPHOSPH9 |
M-phase phosphoprotein 9 |
826 |
0.5 |
| chr7_108060925_108061076 | 0.11 |
NRCAM |
neuronal cell adhesion molecule |
35765 |
0.18 |
| chr4_81018676_81018827 | 0.11 |
ANTXR2 |
anthrax toxin receptor 2 |
24125 |
0.24 |
| chr1_115809791_115809982 | 0.11 |
RP4-663N10.1 |
|
15769 |
0.26 |
| chrY_16636064_16636659 | 0.11 |
NLGN4Y |
neuroligin 4, Y-linked |
93 |
0.99 |
| chr15_29407577_29408042 | 0.11 |
APBA2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
71282 |
0.11 |
| chrX_153978145_153979054 | 0.11 |
GAB3 |
GRB2-associated binding protein 3 |
733 |
0.55 |
| chr8_110551213_110551689 | 0.11 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
489 |
0.82 |
| chr13_24006549_24006700 | 0.11 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
1217 |
0.63 |
| chr3_155053149_155053300 | 0.11 |
ENSG00000272096 |
. |
102388 |
0.07 |
| chr19_17421267_17421418 | 0.11 |
ABHD8 |
abhydrolase domain containing 8 |
297 |
0.69 |
| chr7_24796179_24797014 | 0.11 |
DFNA5 |
deafness, autosomal dominant 5 |
498 |
0.88 |
| chr14_22957215_22957366 | 0.11 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
1119 |
0.33 |
| chr10_14051003_14051266 | 0.11 |
FRMD4A |
FERM domain containing 4A |
602 |
0.78 |
| chr17_76377369_76377979 | 0.11 |
PGS1 |
phosphatidylglycerophosphate synthase 1 |
2915 |
0.19 |
| chr7_27881960_27882111 | 0.11 |
TAX1BP1 |
Tax1 (human T-cell leukemia virus type I) binding protein 1 |
14615 |
0.23 |
| chr9_74516561_74516712 | 0.11 |
ENSG00000200922 |
. |
830 |
0.62 |
| chr10_115235598_115235749 | 0.11 |
HABP2 |
hyaluronan binding protein 2 |
74923 |
0.11 |
| chr11_119235533_119235684 | 0.11 |
USP2 |
ubiquitin specific peptidase 2 |
691 |
0.48 |
| chr7_86842084_86842235 | 0.11 |
ENSG00000200397 |
. |
5869 |
0.19 |
| chr8_22876157_22876423 | 0.11 |
RP11-875O11.1 |
|
558 |
0.66 |
| chr15_82337650_82337919 | 0.11 |
MEX3B |
mex-3 RNA binding family member B |
676 |
0.58 |
| chr11_43600318_43600534 | 0.11 |
ENSG00000199077 |
. |
2518 |
0.32 |
| chr1_226252983_226253134 | 0.11 |
H3F3A |
H3 histone, family 3A |
1380 |
0.39 |
| chr12_26112779_26113361 | 0.11 |
RASSF8-AS1 |
RASSF8 atnisense RNA 1 |
372 |
0.8 |
| chr9_130638533_130638684 | 0.11 |
AK1 |
adenylate kinase 1 |
1322 |
0.23 |
| chr3_16006218_16006514 | 0.11 |
ENSG00000207815 |
. |
91088 |
0.08 |
| chr2_127415770_127416175 | 0.11 |
GYPC |
glycophorin C (Gerbich blood group) |
2212 |
0.42 |
| chr10_102414163_102414356 | 0.11 |
PAX2 |
paired box 2 |
91209 |
0.07 |
| chr18_46474661_46474872 | 0.10 |
SMAD7 |
SMAD family member 7 |
109 |
0.98 |
| chr2_88124610_88124761 | 0.10 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
601 |
0.82 |
| chr1_103574265_103574420 | 0.10 |
COL11A1 |
collagen, type XI, alpha 1 |
290 |
0.95 |
| chr7_28068761_28068999 | 0.10 |
JAZF1 |
JAZF zinc finger 1 |
8462 |
0.28 |
| chr1_202597824_202597975 | 0.10 |
SYT2 |
synaptotagmin II |
14682 |
0.19 |
| chr8_146018268_146018550 | 0.10 |
RPL8 |
ribosomal protein L8 |
437 |
0.71 |
| chr7_93632108_93632388 | 0.10 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
1425 |
0.47 |
| chr15_101136944_101137114 | 0.10 |
LINS |
lines homolog (Drosophila) |
112 |
0.95 |
| chr11_19735220_19735722 | 0.10 |
NAV2 |
neuron navigator 2 |
328 |
0.92 |
| chr9_110512814_110512965 | 0.10 |
AL162389.1 |
Uncharacterized protein |
27530 |
0.19 |
| chr9_113828644_113828795 | 0.10 |
LPAR1 |
lysophosphatidic acid receptor 1 |
27738 |
0.2 |
| chr4_24980238_24980389 | 0.10 |
CCDC149 |
coiled-coil domain containing 149 |
1457 |
0.52 |
| chr15_102286896_102287244 | 0.10 |
RP11-89K11.1 |
Uncharacterized protein |
1157 |
0.38 |
| chr22_28838455_28839140 | 0.10 |
ENSG00000199797 |
. |
139055 |
0.04 |
| chr12_97272342_97272601 | 0.10 |
NEDD1 |
neural precursor cell expressed, developmentally down-regulated 1 |
28530 |
0.25 |
| chr16_9051858_9052009 | 0.10 |
USP7 |
ubiquitin specific peptidase 7 (herpes virus-associated) |
1696 |
0.4 |
| chr1_9477600_9477751 | 0.10 |
ENSG00000252956 |
. |
20162 |
0.21 |
| chr2_131862982_131863218 | 0.10 |
PLEKHB2 |
pleckstrin homology domain containing, family B (evectins) member 2 |
174 |
0.95 |
| chr5_53815643_53816423 | 0.10 |
SNX18 |
sorting nexin 18 |
2440 |
0.4 |
| chr18_21598879_21599226 | 0.10 |
TTC39C |
tetratricopeptide repeat domain 39C |
2454 |
0.25 |
| chr10_74094555_74094706 | 0.10 |
DNAJB12 |
DnaJ (Hsp40) homolog, subfamily B, member 12 |
10089 |
0.19 |
| chr19_53757624_53758099 | 0.10 |
ZNF677 |
zinc finger protein 677 |
80 |
0.94 |
| chr9_139971485_139971648 | 0.10 |
UAP1L1 |
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 |
387 |
0.59 |
| chr9_135547712_135547863 | 0.10 |
DDX31 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
1999 |
0.3 |
| chr11_4658372_4658526 | 0.10 |
OR51D1 |
olfactory receptor, family 51, subfamily D, member 1 |
2496 |
0.19 |
| chr19_56676041_56676192 | 0.10 |
GALP |
galanin-like peptide |
11273 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.2 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0048867 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.0 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0021894 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.5 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.1 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.0 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0007141 | male meiosis I(GO:0007141) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0044447 | axoneme part(GO:0044447) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |