Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
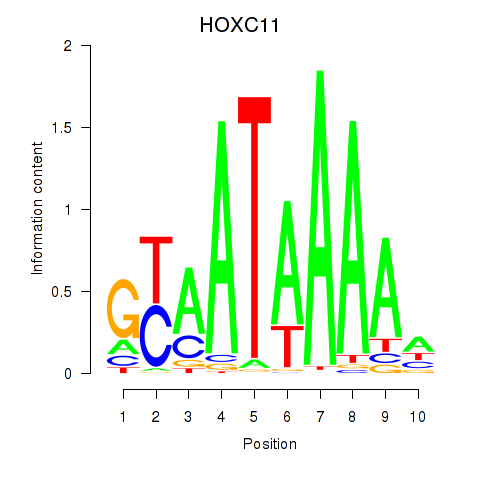
Results for HOXC11
Z-value: 0.22
Transcription factors associated with HOXC11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC11
|
ENSG00000123388.4 | homeobox C11 |
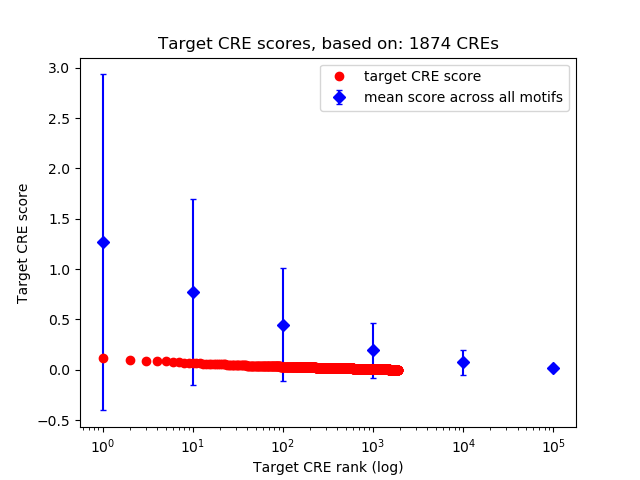
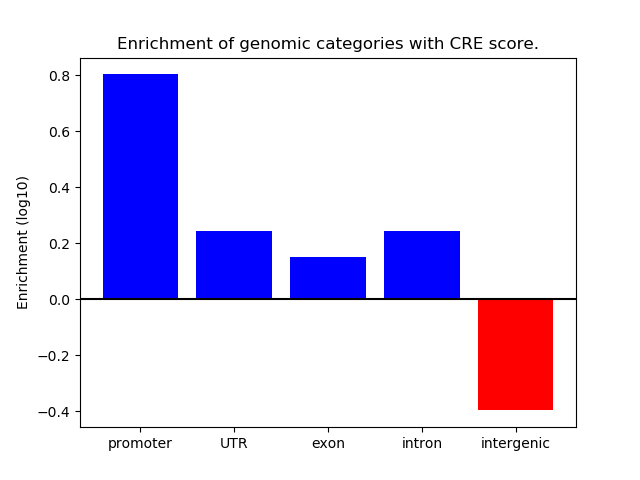
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_54366652_54366803 | HOXC11 | 183 | 0.827388 | 0.31 | 4.1e-01 | Click! |
| chr12_54367124_54367311 | HOXC11 | 305 | 0.691051 | 0.31 | 4.2e-01 | Click! |
| chr12_54366891_54367088 | HOXC11 | 77 | 0.901702 | 0.20 | 6.0e-01 | Click! |
| chr12_54367495_54368083 | HOXC11 | 877 | 0.242171 | -0.19 | 6.2e-01 | Click! |
Activity of the HOXC11 motif across conditions
Conditions sorted by the z-value of the HOXC11 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
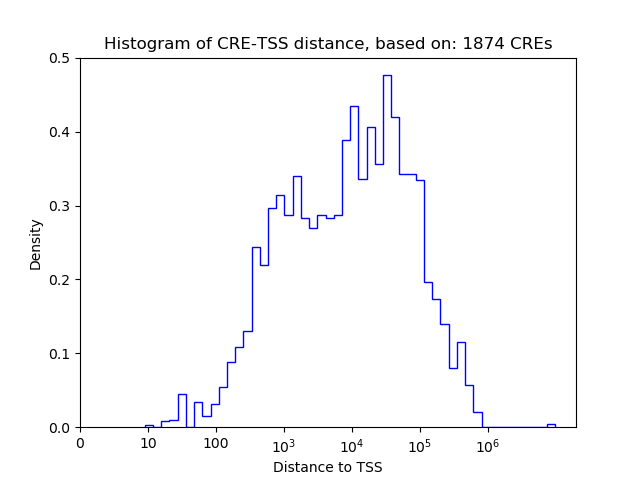
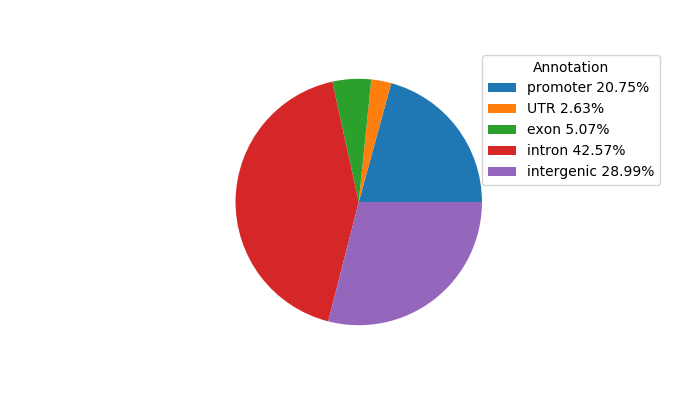
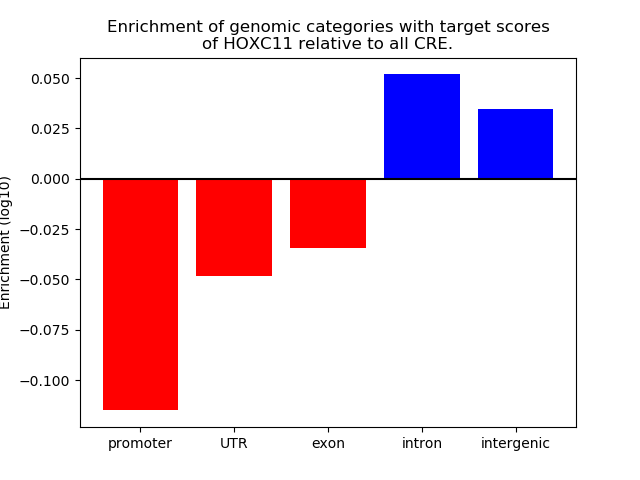
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_183893852_183894003 | 0.11 |
AP2M1 |
adaptor-related protein complex 2, mu 1 subunit |
639 |
0.55 |
| chr7_121514140_121514423 | 0.09 |
PTPRZ1 |
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
866 |
0.74 |
| chr2_213977055_213977206 | 0.09 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
36223 |
0.21 |
| chr7_27138540_27138742 | 0.09 |
HOTAIRM1 |
HOXA transcript antisense RNA, myeloid-specific 1 |
183 |
0.86 |
| chr2_151385576_151386025 | 0.09 |
RND3 |
Rho family GTPase 3 |
9725 |
0.33 |
| chr2_184180506_184180887 | 0.08 |
NUP35 |
nucleoporin 35kDa |
191523 |
0.03 |
| chr6_128529501_128529738 | 0.08 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
113707 |
0.07 |
| chr1_170631765_170631991 | 0.07 |
PRRX1 |
paired related homeobox 1 |
397 |
0.91 |
| chr10_89686715_89686866 | 0.07 |
RP11-380G5.2 |
|
48341 |
0.12 |
| chr12_56308297_56308448 | 0.07 |
WIBG |
within bgcn homolog (Drosophila) |
12524 |
0.09 |
| chr19_4059827_4060036 | 0.07 |
CTD-2622I13.3 |
|
2816 |
0.16 |
| chr4_163496269_163496420 | 0.06 |
FSTL5 |
follistatin-like 5 |
411157 |
0.01 |
| chr3_116755274_116755493 | 0.06 |
ENSG00000200372 |
. |
71309 |
0.12 |
| chr10_104253427_104253578 | 0.06 |
ACTR1A |
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
8933 |
0.1 |
| chr4_39570973_39571124 | 0.06 |
UGDH |
UDP-glucose 6-dehydrogenase |
41117 |
0.11 |
| chr8_83181283_83181434 | 0.06 |
SNX16 |
sorting nexin 16 |
426257 |
0.01 |
| chr8_42063953_42065062 | 0.06 |
PLAT |
plasminogen activator, tissue |
576 |
0.71 |
| chr13_107382919_107383070 | 0.06 |
ARGLU1 |
arginine and glutamate rich 1 |
162482 |
0.04 |
| chr14_81453709_81453897 | 0.06 |
CEP128 |
centrosomal protein 128kDa |
27955 |
0.21 |
| chr11_22380798_22380949 | 0.06 |
CTD-2140G10.4 |
|
2967 |
0.27 |
| chr3_29327656_29328192 | 0.06 |
RBMS3-AS3 |
RBMS3 antisense RNA 3 |
4293 |
0.26 |
| chr8_91782901_91783052 | 0.05 |
NECAB1 |
N-terminal EF-hand calcium binding protein 1 |
20802 |
0.19 |
| chr9_81871537_81871846 | 0.05 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
314997 |
0.01 |
| chr17_32078540_32078804 | 0.05 |
ENSG00000221699 |
. |
122805 |
0.05 |
| chr10_120966033_120966362 | 0.05 |
GRK5 |
G protein-coupled receptor kinase 5 |
904 |
0.45 |
| chrX_31284117_31284268 | 0.05 |
DMD |
dystrophin |
782 |
0.77 |
| chr9_133814218_133814626 | 0.05 |
FIBCD1 |
fibrinogen C domain containing 1 |
33 |
0.97 |
| chr1_178310607_178310919 | 0.05 |
RASAL2 |
RAS protein activator like 2 |
157 |
0.97 |
| chr1_216897072_216897310 | 0.05 |
ESRRG |
estrogen-related receptor gamma |
396 |
0.93 |
| chr5_67483384_67483535 | 0.05 |
RP11-404L6.2 |
Uncharacterized protein |
2245 |
0.34 |
| chr11_94671451_94671602 | 0.05 |
RP11-856F16.2 |
|
12840 |
0.19 |
| chr13_32859694_32859925 | 0.05 |
FRY |
furry homolog (Drosophila) |
2179 |
0.28 |
| chr6_108486676_108487297 | 0.04 |
OSTM1 |
osteopetrosis associated transmembrane protein 1 |
58 |
0.78 |
| chr20_52517333_52517726 | 0.04 |
AC005220.3 |
|
39170 |
0.2 |
| chr20_11882046_11882285 | 0.04 |
BTBD3 |
BTB (POZ) domain containing 3 |
9024 |
0.24 |
| chr3_168890820_168890971 | 0.04 |
MECOM |
MDS1 and EVI1 complex locus |
25373 |
0.27 |
| chr10_24756981_24757214 | 0.04 |
KIAA1217 |
KIAA1217 |
1637 |
0.45 |
| chr5_58412336_58412487 | 0.04 |
RP11-266N13.2 |
|
76823 |
0.11 |
| chr12_93515628_93515779 | 0.04 |
RP11-511B23.2 |
|
17164 |
0.2 |
| chr5_145319076_145319227 | 0.04 |
SH3RF2 |
SH3 domain containing ring finger 2 |
1711 |
0.46 |
| chr12_11325045_11325196 | 0.04 |
RP11-785H5.2 |
|
1168 |
0.4 |
| chr8_8745358_8745509 | 0.04 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
5722 |
0.22 |
| chr9_20511940_20512213 | 0.04 |
ENSG00000264941 |
. |
9736 |
0.23 |
| chr14_85707273_85707789 | 0.04 |
ENSG00000252529 |
. |
30745 |
0.26 |
| chr17_79315981_79316442 | 0.04 |
TMEM105 |
transmembrane protein 105 |
11737 |
0.13 |
| chr12_96184496_96184659 | 0.04 |
NTN4 |
netrin 4 |
35 |
0.97 |
| chr10_83723729_83723880 | 0.04 |
NRG3 |
neuregulin 3 |
86087 |
0.1 |
| chr4_36534650_36534801 | 0.04 |
RP11-431M7.2 |
|
140693 |
0.05 |
| chr5_112822732_112823004 | 0.04 |
MCC |
mutated in colorectal cancers |
1659 |
0.35 |
| chr15_93428286_93428633 | 0.04 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
1933 |
0.32 |
| chr2_198572573_198572724 | 0.04 |
MARS2 |
methionyl-tRNA synthetase 2, mitochondrial |
2561 |
0.27 |
| chr15_25637624_25637775 | 0.04 |
UBE3A |
ubiquitin protein ligase E3A |
12954 |
0.11 |
| chr3_15823018_15823169 | 0.04 |
ANKRD28 |
ankyrin repeat domain 28 |
14935 |
0.19 |
| chr1_70768783_70768934 | 0.04 |
ANKRD13C |
ankyrin repeat domain 13C |
51547 |
0.11 |
| chr2_222287657_222287808 | 0.04 |
EPHA4 |
EPH receptor A4 |
79548 |
0.11 |
| chr4_153269690_153269947 | 0.04 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
140 |
0.97 |
| chr11_79150724_79151047 | 0.04 |
TENM4 |
teneurin transmembrane protein 4 |
810 |
0.68 |
| chr17_62971851_62972399 | 0.04 |
AMZ2P1 |
archaelysin family metallopeptidase 2 pseudogene 1 |
2490 |
0.23 |
| chr10_22899285_22899436 | 0.04 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
18718 |
0.27 |
| chr8_49321315_49321611 | 0.04 |
ENSG00000252710 |
. |
100873 |
0.08 |
| chr9_12859535_12860149 | 0.04 |
ENSG00000222658 |
. |
25478 |
0.2 |
| chr4_26240703_26241213 | 0.04 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
33271 |
0.24 |
| chr3_49538097_49538499 | 0.04 |
DAG1 |
dystroglycan 1 (dystrophin-associated glycoprotein 1) |
30546 |
0.11 |
| chr12_86228304_86228474 | 0.04 |
RASSF9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
1959 |
0.44 |
| chr15_48739773_48739924 | 0.04 |
DUT |
deoxyuridine triphosphatase |
115245 |
0.05 |
| chr6_113966553_113966868 | 0.04 |
ENSG00000266650 |
. |
42593 |
0.17 |
| chr1_115711115_115711431 | 0.04 |
TSPAN2 |
tetraspanin 2 |
79152 |
0.1 |
| chr11_77072652_77072803 | 0.04 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
12395 |
0.24 |
| chr6_153911669_153911820 | 0.04 |
ENSG00000199246 |
. |
90838 |
0.09 |
| chr3_151743926_151744077 | 0.04 |
RP11-454C18.2 |
|
98038 |
0.08 |
| chr5_57751342_57751690 | 0.04 |
PLK2 |
polo-like kinase 2 |
4571 |
0.26 |
| chr13_43649218_43649369 | 0.04 |
DNAJC15 |
DnaJ (Hsp40) homolog, subfamily C, member 15 |
51954 |
0.17 |
| chr14_77998583_77998734 | 0.04 |
ENSG00000240233 |
. |
15554 |
0.15 |
| chr13_80511227_80511398 | 0.03 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
402482 |
0.01 |
| chr2_166477820_166478090 | 0.03 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
47336 |
0.18 |
| chr9_101891255_101891406 | 0.03 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
1114 |
0.52 |
| chr9_12775321_12775833 | 0.03 |
LURAP1L |
leucine rich adaptor protein 1-like |
557 |
0.78 |
| chr1_224690897_224691172 | 0.03 |
WDR26 |
WD repeat domain 26 |
66299 |
0.1 |
| chr6_167539321_167539472 | 0.03 |
CCR6 |
chemokine (C-C motif) receptor 6 |
3137 |
0.25 |
| chr9_100173959_100174113 | 0.03 |
TDRD7 |
tudor domain containing 7 |
196 |
0.95 |
| chr6_53928552_53928746 | 0.03 |
MLIP-AS1 |
MLIP antisense RNA 1 |
15875 |
0.2 |
| chr2_121716938_121717089 | 0.03 |
ENSG00000221321 |
. |
75636 |
0.11 |
| chr17_55131735_55131912 | 0.03 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
30630 |
0.13 |
| chrX_15965760_15965911 | 0.03 |
ENSG00000200566 |
. |
31409 |
0.2 |
| chr3_104994911_104995062 | 0.03 |
ALCAM |
activated leukocyte cell adhesion molecule |
90767 |
0.1 |
| chr1_83031111_83031295 | 0.03 |
LPHN2 |
latrophilin 2 |
585630 |
0.0 |
| chr10_4174092_4174250 | 0.03 |
KLF6 |
Kruppel-like factor 6 |
346698 |
0.01 |
| chr10_33620428_33620906 | 0.03 |
NRP1 |
neuropilin 1 |
2643 |
0.37 |
| chr6_56598112_56598263 | 0.03 |
DST |
dystonin |
52490 |
0.16 |
| chr5_143300598_143300749 | 0.03 |
HMHB1 |
histocompatibility (minor) HB-1 |
108947 |
0.07 |
| chr14_89507425_89507846 | 0.03 |
FOXN3 |
forkhead box N3 |
139453 |
0.05 |
| chr10_62174410_62174561 | 0.03 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
24997 |
0.28 |
| chr12_95606332_95606483 | 0.03 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
4748 |
0.2 |
| chr5_180676019_180676390 | 0.03 |
GNB2L1 |
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
1108 |
0.22 |
| chr22_25507961_25508112 | 0.03 |
CTA-221G9.11 |
|
623 |
0.77 |
| chr11_111381371_111381522 | 0.03 |
BTG4 |
B-cell translocation gene 4 |
1618 |
0.2 |
| chr6_3501213_3501364 | 0.03 |
SLC22A23 |
solute carrier family 22, member 23 |
44032 |
0.18 |
| chr8_97583871_97584022 | 0.03 |
SDC2 |
syndecan 2 |
8631 |
0.28 |
| chr2_213976639_213976790 | 0.03 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
36639 |
0.21 |
| chr2_134432207_134432889 | 0.03 |
ENSG00000200708 |
. |
78886 |
0.11 |
| chr5_32164159_32164310 | 0.03 |
GOLPH3 |
golgi phosphoprotein 3 (coat-protein) |
10222 |
0.19 |
| chr16_81577563_81578015 | 0.03 |
CMIP |
c-Maf inducing protein |
48835 |
0.17 |
| chr5_110846365_110846666 | 0.03 |
STARD4-AS1 |
STARD4 antisense RNA 1 |
1409 |
0.37 |
| chr19_4717454_4717717 | 0.03 |
DPP9 |
dipeptidyl-peptidase 9 |
315 |
0.83 |
| chr22_36291410_36291691 | 0.03 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
43395 |
0.19 |
| chr4_139160727_139161292 | 0.03 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
2494 |
0.42 |
| chr15_69367071_69367289 | 0.03 |
RP11-809H16.4 |
|
31275 |
0.17 |
| chr5_124748322_124748473 | 0.03 |
ENSG00000222107 |
. |
61573 |
0.16 |
| chr15_26273691_26273918 | 0.03 |
ENSG00000212604 |
. |
19583 |
0.26 |
| chr6_111947186_111947610 | 0.03 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
19917 |
0.16 |
| chr12_123210436_123210665 | 0.03 |
HCAR1 |
hydroxycarboxylic acid receptor 1 |
4579 |
0.15 |
| chr10_131323764_131323955 | 0.03 |
AL355531.2 |
Uncharacterized protein |
14334 |
0.23 |
| chr18_7958694_7958932 | 0.03 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
11861 |
0.23 |
| chrX_151140998_151141149 | 0.03 |
GABRE |
gamma-aminobutyric acid (GABA) A receptor, epsilon |
2070 |
0.24 |
| chr15_38361508_38361698 | 0.03 |
TMCO5A |
transmembrane and coiled-coil domains 5A |
134145 |
0.05 |
| chr12_80329810_80330005 | 0.03 |
PPP1R12A |
protein phosphatase 1, regulatory subunit 12A |
667 |
0.6 |
| chr1_115820493_115820644 | 0.03 |
RP4-663N10.1 |
|
5087 |
0.3 |
| chr3_37031282_37031460 | 0.03 |
EPM2AIP1 |
EPM2A (laforin) interacting protein 1 |
3424 |
0.2 |
| chr16_68299633_68299784 | 0.03 |
SLC7A6 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
289 |
0.77 |
| chr1_94789725_94789884 | 0.03 |
ARHGAP29 |
Rho GTPase activating protein 29 |
86615 |
0.08 |
| chr1_172327726_172327959 | 0.03 |
ENSG00000206684 |
. |
7838 |
0.18 |
| chr4_40059743_40060131 | 0.03 |
N4BP2 |
NEDD4 binding protein 2 |
1381 |
0.41 |
| chr11_89113858_89114276 | 0.03 |
NOX4 |
NADPH oxidase 4 |
109833 |
0.07 |
| chr16_80181289_80181449 | 0.03 |
DYNLRB2 |
dynein, light chain, roadblock-type 2 |
393262 |
0.01 |
| chr17_55372937_55373088 | 0.03 |
MSI2 |
musashi RNA-binding protein 2 |
966 |
0.68 |
| chr4_85420275_85420452 | 0.03 |
NKX6-1 |
NK6 homeobox 1 |
760 |
0.77 |
| chr16_73089798_73090050 | 0.03 |
ZFHX3 |
zinc finger homeobox 3 |
3673 |
0.28 |
| chr1_218524044_218525060 | 0.03 |
TGFB2 |
transforming growth factor, beta 2 |
4975 |
0.24 |
| chr18_20612011_20612377 | 0.03 |
ENSG00000223023 |
. |
7635 |
0.17 |
| chr11_82904254_82904445 | 0.03 |
ANKRD42 |
ankyrin repeat domain 42 |
432 |
0.8 |
| chr8_103664786_103665058 | 0.03 |
KLF10 |
Kruppel-like factor 10 |
1681 |
0.44 |
| chr21_16429371_16429522 | 0.03 |
NRIP1 |
nuclear receptor interacting protein 1 |
7680 |
0.28 |
| chr14_23448905_23449056 | 0.03 |
AJUBA |
ajuba LIM protein |
1815 |
0.14 |
| chr4_169752801_169753072 | 0.03 |
PALLD |
palladin, cytoskeletal associated protein |
220 |
0.9 |
| chr2_27861313_27861464 | 0.03 |
ENSG00000206731 |
. |
3648 |
0.13 |
| chrX_78666635_78666786 | 0.03 |
ITM2A |
integral membrane protein 2A |
43661 |
0.22 |
| chr16_48641989_48642140 | 0.03 |
N4BP1 |
NEDD4 binding protein 1 |
2056 |
0.33 |
| chr6_125401116_125401267 | 0.03 |
RNF217 |
ring finger protein 217 |
31833 |
0.2 |
| chr18_19753166_19753317 | 0.03 |
GATA6 |
GATA binding protein 6 |
2347 |
0.21 |
| chr2_85196569_85196904 | 0.03 |
KCMF1 |
potassium channel modulatory factor 1 |
1480 |
0.46 |
| chr2_235167151_235167323 | 0.03 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
207806 |
0.02 |
| chr3_114225259_114225410 | 0.03 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
51670 |
0.16 |
| chr1_159931930_159932191 | 0.03 |
SLAMF9 |
SLAM family member 9 |
8016 |
0.12 |
| chr5_53386895_53387046 | 0.03 |
ENSG00000265421 |
. |
15557 |
0.24 |
| chr3_191048468_191048928 | 0.03 |
UTS2B |
urotensin 2B |
373 |
0.85 |
| chr10_116634887_116635113 | 0.03 |
FAM160B1 |
family with sequence similarity 160, member B1 |
14450 |
0.25 |
| chr17_814224_814439 | 0.03 |
RP11-676J12.7 |
Uncharacterized protein |
1459 |
0.38 |
| chr6_17914331_17914482 | 0.03 |
KIF13A |
kinesin family member 13A |
73288 |
0.11 |
| chr22_24114508_24114741 | 0.03 |
MMP11 |
matrix metallopeptidase 11 (stromelysin 3) |
382 |
0.71 |
| chr17_4135715_4135866 | 0.03 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
31352 |
0.12 |
| chr1_241271770_241271921 | 0.03 |
ENSG00000265831 |
. |
23727 |
0.23 |
| chr13_21654404_21654812 | 0.03 |
LATS2 |
large tumor suppressor kinase 2 |
18922 |
0.14 |
| chr3_81724848_81724999 | 0.03 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
67857 |
0.14 |
| chr3_64149261_64149412 | 0.03 |
PRICKLE2-AS3 |
PRICKLE2 antisense RNA 3 |
23884 |
0.16 |
| chr4_114367456_114367607 | 0.03 |
ENSG00000206820 |
. |
26052 |
0.18 |
| chr8_34689780_34689931 | 0.03 |
ENSG00000265560 |
. |
137869 |
0.05 |
| chr13_20844899_20845050 | 0.03 |
GJB6 |
gap junction protein, beta 6, 30kDa |
38440 |
0.17 |
| chr11_11472823_11472974 | 0.03 |
CSNK2A3 |
casein kinase 2, alpha 3 polypeptide |
97994 |
0.07 |
| chr11_89955284_89955841 | 0.03 |
CHORDC1 |
cysteine and histidine-rich domain (CHORD) containing 1 |
665 |
0.79 |
| chr18_55889348_55889572 | 0.03 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
657 |
0.77 |
| chr6_17413520_17413671 | 0.03 |
CAP2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
19652 |
0.26 |
| chr5_60728941_60729092 | 0.03 |
ZSWIM6 |
zinc finger, SWIM-type containing 6 |
100916 |
0.08 |
| chr7_141628433_141628584 | 0.03 |
OR9A4 |
olfactory receptor, family 9, subfamily A, member 4 |
9891 |
0.12 |
| chr15_99788624_99789059 | 0.03 |
TTC23 |
tetratricopeptide repeat domain 23 |
895 |
0.51 |
| chr16_80603632_80603783 | 0.03 |
RP11-525K10.3 |
|
6675 |
0.18 |
| chr2_166802065_166802216 | 0.03 |
TTC21B |
tetratricopeptide repeat domain 21B |
8213 |
0.19 |
| chr6_21417907_21418058 | 0.03 |
SOX4 |
SRY (sex determining region Y)-box 4 |
175990 |
0.04 |
| chrX_119596476_119596627 | 0.03 |
LAMP2 |
lysosomal-associated membrane protein 2 |
6296 |
0.25 |
| chr8_94859053_94859497 | 0.03 |
TMEM67 |
transmembrane protein 67 |
61778 |
0.09 |
| chr20_6764834_6765429 | 0.03 |
BMP2 |
bone morphogenetic protein 2 |
16820 |
0.28 |
| chr13_44389557_44389806 | 0.03 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
28637 |
0.21 |
| chr13_30002091_30002575 | 0.03 |
MTUS2 |
microtubule associated tumor suppressor candidate 2 |
431 |
0.89 |
| chr3_39448821_39449415 | 0.03 |
ENSG00000206760 |
. |
762 |
0.4 |
| chr7_25896786_25897110 | 0.03 |
ENSG00000199085 |
. |
92658 |
0.09 |
| chr17_57699008_57699159 | 0.03 |
CLTC |
clathrin, heavy chain (Hc) |
1790 |
0.37 |
| chr6_54564223_54564374 | 0.02 |
ENSG00000251946 |
. |
86887 |
0.1 |
| chr4_83787091_83787242 | 0.02 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
918 |
0.55 |
| chr13_99629621_99629906 | 0.02 |
DOCK9 |
dedicator of cytokinesis 9 |
481 |
0.85 |
| chr5_114868008_114868159 | 0.02 |
FEM1C |
fem-1 homolog c (C. elegans) |
12508 |
0.22 |
| chr19_45995836_45996758 | 0.02 |
RTN2 |
reticulon 2 |
228 |
0.87 |
| chr4_14728626_14728777 | 0.02 |
ENSG00000202449 |
. |
36367 |
0.24 |
| chrX_73068432_73068583 | 0.02 |
RP13-216E22.5 |
|
120813 |
0.06 |
| chr14_45367367_45367627 | 0.02 |
C14orf28 |
chromosome 14 open reading frame 28 |
959 |
0.41 |
| chr11_27486888_27487039 | 0.02 |
RP11-159H22.2 |
|
6313 |
0.18 |
| chr3_160700419_160700570 | 0.02 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
118461 |
0.06 |
| chr15_71054590_71054766 | 0.02 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
1091 |
0.55 |
| chr9_132175724_132175951 | 0.02 |
ENSG00000242281 |
. |
43097 |
0.14 |
| chr16_20922292_20922443 | 0.02 |
LYRM1 |
LYR motif containing 1 |
9941 |
0.16 |
| chr11_94441338_94441489 | 0.02 |
AMOTL1 |
angiomotin like 1 |
1816 |
0.4 |
| chr17_62648329_62648480 | 0.02 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
9626 |
0.22 |
| chr8_10268256_10268407 | 0.02 |
RP11-981G7.1 |
|
22851 |
0.2 |
| chr10_8315388_8315539 | 0.02 |
GATA3 |
GATA binding protein 3 |
218694 |
0.02 |
| chr16_50400842_50400993 | 0.02 |
RP11-21B23.1 |
|
1574 |
0.3 |
| chr7_17977598_17977749 | 0.02 |
SNX13 |
sorting nexin 13 |
2418 |
0.38 |
| chr3_120168336_120168615 | 0.02 |
FSTL1 |
follistatin-like 1 |
1363 |
0.54 |
| chr1_23476622_23476773 | 0.02 |
ENSG00000206935 |
. |
12500 |
0.13 |
| chr5_128796123_128796371 | 0.02 |
ADAMTS19-AS1 |
ADAMTS19 antisense RNA 1 |
135 |
0.71 |
| chr18_21418407_21418558 | 0.02 |
LAMA3 |
laminin, alpha 3 |
4565 |
0.28 |
| chr14_96681269_96681420 | 0.02 |
RP11-404P21.5 |
|
4147 |
0.14 |
| chr12_64480628_64481033 | 0.02 |
RP11-196H14.2 |
|
10568 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |