Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
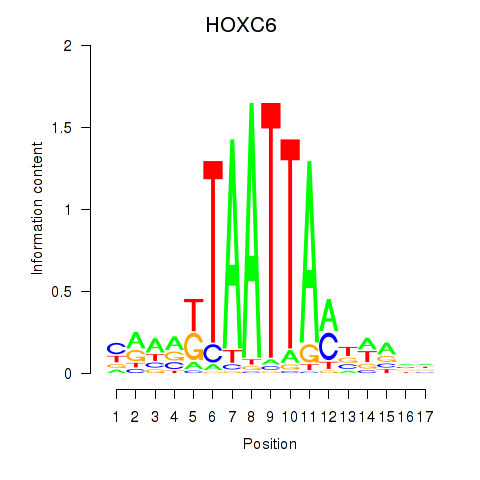
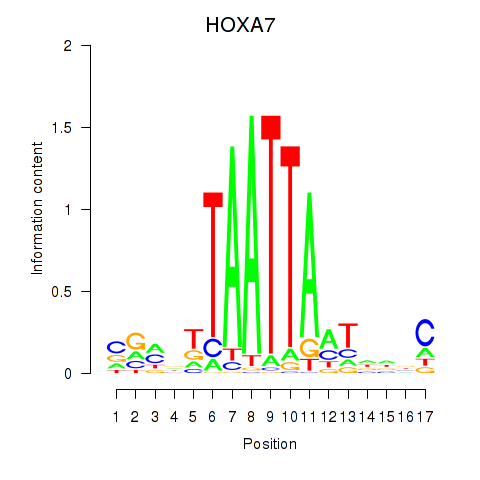
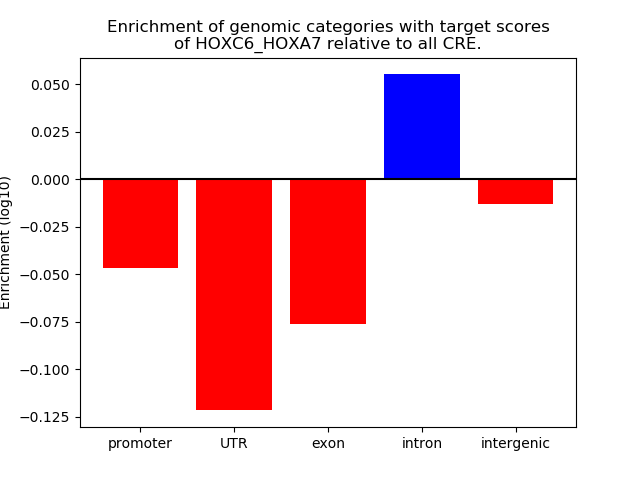
Results for HOXC6_HOXA7
Z-value: 0.64
Transcription factors associated with HOXC6_HOXA7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC6
|
ENSG00000197757.7 | homeobox C6 |
|
HOXA7
|
ENSG00000122592.6 | homeobox A7 |
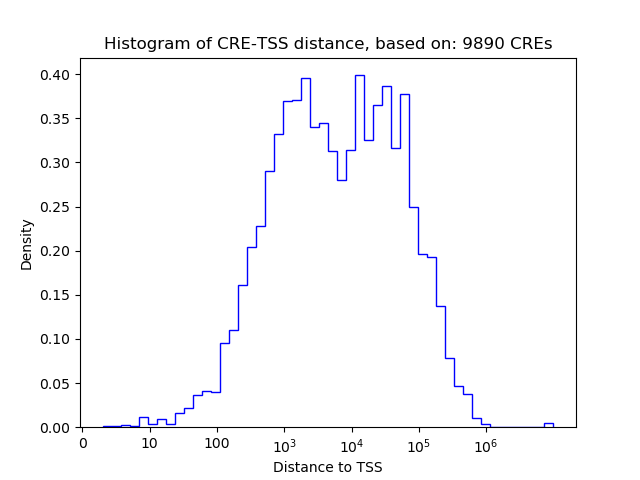
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_27199352_27199503 | HOXA7 | 1872 | 0.117379 | -0.76 | 1.9e-02 | Click! |
| chr7_27197659_27197810 | HOXA7 | 179 | 0.830311 | -0.51 | 1.6e-01 | Click! |
| chr7_27198059_27198210 | HOXA7 | 579 | 0.443014 | -0.48 | 1.9e-01 | Click! |
| chr7_27196933_27197228 | HOXA7 | 475 | 0.528812 | -0.47 | 2.0e-01 | Click! |
| chr7_27198968_27199186 | HOXA7 | 1522 | 0.146054 | -0.44 | 2.4e-01 | Click! |
| chr12_54406961_54407131 | HOXC6 | 2635 | 0.091558 | 0.61 | 8.4e-02 | Click! |
| chr12_54420892_54421043 | HOXC6 | 1175 | 0.224136 | 0.47 | 2.0e-01 | Click! |
| chr12_54408009_54408173 | HOXC6 | 1590 | 0.143819 | 0.46 | 2.1e-01 | Click! |
| chr12_54408811_54408962 | HOXC6 | 795 | 0.316110 | 0.45 | 2.2e-01 | Click! |
| chr12_54384773_54384996 | HOXC6 | 476 | 0.404093 | 0.44 | 2.4e-01 | Click! |
Activity of the HOXC6_HOXA7 motif across conditions
Conditions sorted by the z-value of the HOXC6_HOXA7 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
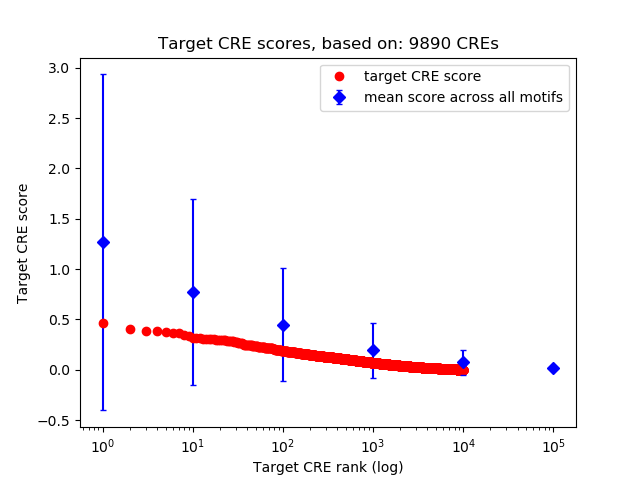
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_22968352_22968523 | 0.46 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
12266 |
0.1 |
| chrX_19815653_19816006 | 0.40 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
2040 |
0.46 |
| chr5_81762775_81762926 | 0.39 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
161684 |
0.04 |
| chr14_22947593_22947744 | 0.39 |
TRAJ60 |
T cell receptor alpha joining 60 (pseudogene) |
2372 |
0.15 |
| chr3_124303613_124304383 | 0.38 |
KALRN |
kalirin, RhoGEF kinase |
329 |
0.93 |
| chr19_50062782_50062933 | 0.37 |
NOSIP |
nitric oxide synthase interacting protein |
1030 |
0.27 |
| chr18_56106091_56106384 | 0.37 |
ENSG00000207778 |
. |
12069 |
0.17 |
| chr5_66646304_66646752 | 0.34 |
ENSG00000222939 |
. |
113908 |
0.07 |
| chr10_47640402_47640757 | 0.33 |
ANTXRLP1 |
anthrax toxin receptor-like pseudogene 1 |
4212 |
0.25 |
| chr5_35863316_35863652 | 0.32 |
IL7R |
interleukin 7 receptor |
6490 |
0.21 |
| chr20_53217986_53218137 | 0.32 |
DOK5 |
docking protein 5 |
125804 |
0.06 |
| chr6_116834038_116834189 | 0.31 |
TRAPPC3L |
trafficking protein particle complex 3-like |
607 |
0.55 |
| chr15_26095105_26095679 | 0.31 |
ENSG00000266517 |
. |
1420 |
0.42 |
| chr12_27472599_27472750 | 0.31 |
ARNTL2 |
aryl hydrocarbon receptor nuclear translocator-like 2 |
13113 |
0.22 |
| chr4_82327183_82327334 | 0.31 |
RASGEF1B |
RasGEF domain family, member 1B |
65132 |
0.13 |
| chr5_119523820_119523971 | 0.30 |
ENSG00000251975 |
. |
149453 |
0.05 |
| chr12_47611625_47611801 | 0.30 |
PCED1B |
PC-esterase domain containing 1B |
1332 |
0.49 |
| chr3_114136122_114136315 | 0.30 |
ZBTB20-AS1 |
ZBTB20 antisense RNA 1 |
29702 |
0.18 |
| chr7_28429769_28429920 | 0.30 |
CREB5 |
cAMP responsive element binding protein 5 |
19162 |
0.28 |
| chr5_44684027_44684178 | 0.30 |
ENSG00000263556 |
. |
32190 |
0.23 |
| chr17_47766049_47766200 | 0.29 |
SPOP |
speckle-type POZ protein |
10528 |
0.14 |
| chr2_175710954_175711206 | 0.29 |
CHN1 |
chimerin 1 |
53 |
0.98 |
| chr6_145693454_145693782 | 0.29 |
ENSG00000221796 |
. |
23724 |
0.26 |
| chr12_29696081_29696326 | 0.29 |
RP11-310I24.1 |
|
23539 |
0.2 |
| chr14_22985583_22985743 | 0.29 |
TRAJ15 |
T cell receptor alpha joining 15 |
12917 |
0.1 |
| chr3_36504651_36504802 | 0.29 |
STAC |
SH3 and cysteine rich domain |
82661 |
0.11 |
| chr17_37774592_37774840 | 0.29 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
8277 |
0.12 |
| chr4_2486639_2486842 | 0.28 |
RNF4 |
ring finger protein 4 |
4798 |
0.2 |
| chr1_89594453_89594640 | 0.28 |
GBP2 |
guanylate binding protein 2, interferon-inducible |
2749 |
0.27 |
| chr3_186745077_186745229 | 0.28 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
1882 |
0.42 |
| chr14_98603340_98603491 | 0.28 |
C14orf64 |
chromosome 14 open reading frame 64 |
158954 |
0.04 |
| chr12_7985471_7986071 | 0.27 |
SLC2A14 |
solute carrier family 2 (facilitated glucose transporter), member 14 |
359 |
0.8 |
| chr2_38894094_38894271 | 0.27 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
923 |
0.55 |
| chr8_126960676_126960849 | 0.26 |
ENSG00000206695 |
. |
47567 |
0.2 |
| chr9_95730621_95730837 | 0.26 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
4486 |
0.25 |
| chr14_65450405_65450576 | 0.26 |
FNTB |
farnesyltransferase, CAAX box, beta |
2948 |
0.17 |
| chr11_88087417_88087693 | 0.25 |
CTSC |
cathepsin C |
16600 |
0.28 |
| chrX_135732358_135732618 | 0.25 |
CD40LG |
CD40 ligand |
2102 |
0.29 |
| chr5_140457403_140457554 | 0.25 |
PCDHB2 |
protocadherin beta 2 |
16749 |
0.09 |
| chr6_56715487_56715638 | 0.25 |
DST |
dystonin |
1187 |
0.53 |
| chr7_15340100_15340251 | 0.24 |
AGMO |
alkylglycerol monooxygenase |
65637 |
0.15 |
| chr14_100535454_100535614 | 0.24 |
EVL |
Enah/Vasp-like |
2760 |
0.21 |
| chr16_86638626_86639076 | 0.24 |
FOXL1 |
forkhead box L1 |
26736 |
0.16 |
| chr12_12658847_12659363 | 0.24 |
DUSP16 |
dual specificity phosphatase 16 |
14954 |
0.24 |
| chr5_154033985_154034136 | 0.24 |
ENSG00000221552 |
. |
31276 |
0.14 |
| chr3_194409728_194410184 | 0.24 |
FAM43A |
family with sequence similarity 43, member A |
3334 |
0.19 |
| chr14_68774448_68774599 | 0.24 |
ENSG00000243546 |
. |
71267 |
0.11 |
| chr10_6627562_6627792 | 0.24 |
PRKCQ |
protein kinase C, theta |
5414 |
0.34 |
| chr15_58702506_58703018 | 0.24 |
LIPC |
lipase, hepatic |
6 |
0.98 |
| chr7_110732102_110732377 | 0.23 |
LRRN3 |
leucine rich repeat neuronal 3 |
1140 |
0.59 |
| chr5_67576888_67577103 | 0.23 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
860 |
0.74 |
| chr3_98619475_98619750 | 0.23 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
403 |
0.8 |
| chr13_45272325_45272476 | 0.23 |
ENSG00000238932 |
. |
69591 |
0.12 |
| chr1_109751420_109751576 | 0.23 |
ENSG00000238310 |
. |
1028 |
0.46 |
| chr6_11394166_11394363 | 0.23 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
11683 |
0.27 |
| chr6_21004300_21004557 | 0.23 |
ENSG00000252046 |
. |
10629 |
0.32 |
| chr10_17707589_17707769 | 0.23 |
ENSG00000251959 |
. |
13508 |
0.15 |
| chr5_174487191_174487429 | 0.22 |
ENSG00000266890 |
. |
308573 |
0.01 |
| chr20_18446613_18446764 | 0.22 |
DZANK1 |
double zinc ribbon and ankyrin repeat domains 1 |
663 |
0.48 |
| chr1_20649271_20649434 | 0.22 |
RP4-745E8.2 |
|
328 |
0.9 |
| chr3_143445457_143445608 | 0.22 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
121768 |
0.06 |
| chr1_193152081_193152232 | 0.22 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
3628 |
0.25 |
| chrY_15815919_15816489 | 0.22 |
TMSB4Y |
thymosin beta 4, Y-linked |
757 |
0.72 |
| chr15_40112127_40112278 | 0.22 |
RP11-37C7.2 |
|
4572 |
0.19 |
| chr16_12282412_12282563 | 0.22 |
SNX29 |
sorting nexin 29 |
58933 |
0.11 |
| chr15_70867504_70867655 | 0.22 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
127041 |
0.06 |
| chr9_123686452_123686718 | 0.22 |
TRAF1 |
TNF receptor-associated factor 1 |
4462 |
0.25 |
| chr14_57274863_57275150 | 0.21 |
OTX2 |
orthodenticle homeobox 2 |
2160 |
0.26 |
| chr3_189766902_189767149 | 0.21 |
ENSG00000265045 |
. |
64698 |
0.1 |
| chr16_4162545_4162832 | 0.21 |
ADCY9 |
adenylate cyclase 9 |
1339 |
0.51 |
| chr22_34248307_34248458 | 0.21 |
LARGE |
like-glycosyltransferase |
9230 |
0.27 |
| chr18_32626039_32626502 | 0.21 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
4656 |
0.33 |
| chr7_15722890_15723041 | 0.21 |
MEOX2 |
mesenchyme homeobox 2 |
3472 |
0.3 |
| chr1_245551646_245551892 | 0.21 |
ENSG00000221165 |
. |
18133 |
0.26 |
| chr17_21935353_21935504 | 0.21 |
MTRNR2L1 |
MT-RNR2-like 1 |
87009 |
0.09 |
| chr5_60704297_60704575 | 0.21 |
ZSWIM6 |
zinc finger, SWIM-type containing 6 |
76336 |
0.11 |
| chr10_17151338_17151489 | 0.21 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
18423 |
0.23 |
| chr17_66700653_66700804 | 0.21 |
ENSG00000263690 |
. |
61972 |
0.13 |
| chr6_16135670_16135821 | 0.21 |
ENSG00000263712 |
. |
6042 |
0.19 |
| chr3_108549304_108549455 | 0.21 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
7760 |
0.27 |
| chr14_62170526_62170677 | 0.20 |
HIF1A |
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
6261 |
0.26 |
| chr4_157594550_157594701 | 0.20 |
RP11-171N4.3 |
|
30893 |
0.2 |
| chr4_1514296_1514447 | 0.20 |
NKX1-1 |
NK1 homeobox 1 |
114252 |
0.05 |
| chr10_33241565_33241796 | 0.20 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
2911 |
0.37 |
| chr6_16420893_16421044 | 0.20 |
ENSG00000265642 |
. |
7786 |
0.31 |
| chr9_98188423_98189086 | 0.20 |
PTCH1 |
patched 1 |
54013 |
0.12 |
| chr9_88206707_88206867 | 0.20 |
AGTPBP1 |
ATP/GTP binding protein 1 |
89414 |
0.1 |
| chr8_13346130_13346281 | 0.20 |
DLC1 |
deleted in liver cancer 1 |
10545 |
0.24 |
| chr12_111016409_111016682 | 0.19 |
PPTC7 |
PTC7 protein phosphatase homolog (S. cerevisiae) |
4580 |
0.18 |
| chr1_172109378_172109611 | 0.19 |
ENSG00000207949 |
. |
1447 |
0.39 |
| chr10_63813819_63813970 | 0.19 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
4924 |
0.3 |
| chr2_133058367_133058560 | 0.19 |
ENSG00000221288 |
. |
43810 |
0.15 |
| chr1_227501319_227501470 | 0.19 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
3489 |
0.36 |
| chr5_125036350_125036501 | 0.19 |
ENSG00000222107 |
. |
349601 |
0.01 |
| chr13_24146066_24146217 | 0.19 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
1338 |
0.59 |
| chr10_28336370_28336521 | 0.19 |
ARMC4 |
armadillo repeat containing 4 |
48468 |
0.18 |
| chr7_24768620_24768771 | 0.19 |
DFNA5 |
deafness, autosomal dominant 5 |
9980 |
0.29 |
| chr7_28002200_28002423 | 0.19 |
JAZF1 |
JAZF zinc finger 1 |
29364 |
0.22 |
| chr5_39186752_39186903 | 0.19 |
FYB |
FYN binding protein |
16302 |
0.26 |
| chr1_158903283_158903483 | 0.19 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
2025 |
0.36 |
| chr5_67580506_67580669 | 0.19 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
3609 |
0.36 |
| chr12_92534627_92535592 | 0.19 |
C12orf79 |
chromosome 12 open reading frame 79 |
1335 |
0.39 |
| chrX_57618068_57618312 | 0.19 |
ZXDB |
zinc finger, X-linked, duplicated B |
79 |
0.99 |
| chr10_29908483_29908634 | 0.19 |
SVIL |
supervillin |
15343 |
0.18 |
| chrX_72782625_72782944 | 0.18 |
CHIC1 |
cysteine-rich hydrophobic domain 1 |
252 |
0.96 |
| chr11_108040783_108041103 | 0.18 |
NPAT |
nuclear protein, ataxia-telangiectasia locus |
8051 |
0.17 |
| chr14_29254002_29254283 | 0.18 |
RP11-966I7.2 |
|
851 |
0.56 |
| chr2_25500266_25500584 | 0.18 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
25245 |
0.19 |
| chr14_71454929_71455080 | 0.18 |
PCNX |
pecanex homolog (Drosophila) |
24744 |
0.26 |
| chr12_866633_866784 | 0.18 |
WNK1 |
WNK lysine deficient protein kinase 1 |
3976 |
0.25 |
| chr21_35306376_35306527 | 0.18 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
2933 |
0.21 |
| chr4_169564516_169564687 | 0.18 |
RP11-610J23.1 |
|
5137 |
0.22 |
| chr7_39170814_39170965 | 0.18 |
POU6F2 |
POU class 6 homeobox 2 |
45428 |
0.18 |
| chr2_225809390_225809894 | 0.18 |
DOCK10 |
dedicator of cytokinesis 10 |
2140 |
0.45 |
| chr8_77592906_77593199 | 0.18 |
ZFHX4 |
zinc finger homeobox 4 |
402 |
0.86 |
| chr9_124921339_124921603 | 0.18 |
NDUFA8 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8, 19kDa |
595 |
0.48 |
| chr3_105295301_105295452 | 0.18 |
ALCAM |
activated leukocyte cell adhesion molecule |
42930 |
0.21 |
| chrX_57022329_57022623 | 0.18 |
SPIN3 |
spindlin family, member 3 |
507 |
0.84 |
| chr15_60883034_60883482 | 0.18 |
RORA |
RAR-related orphan receptor A |
1482 |
0.47 |
| chr1_85247835_85247986 | 0.18 |
ENSG00000251899 |
. |
10972 |
0.25 |
| chr8_95973021_95973268 | 0.18 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
2908 |
0.2 |
| chr15_58920533_58921016 | 0.18 |
ENSG00000239100 |
. |
28340 |
0.16 |
| chr18_34914843_34915294 | 0.18 |
RP11-797E24.3 |
|
60645 |
0.13 |
| chr17_1992653_1992959 | 0.18 |
RP11-667K14.5 |
|
169 |
0.91 |
| chr1_51447205_51447356 | 0.18 |
CDKN2C |
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
11664 |
0.2 |
| chr8_91554264_91554415 | 0.18 |
ENSG00000199354 |
. |
30983 |
0.2 |
| chr13_39610123_39610274 | 0.18 |
PROSER1 |
proline and serine rich 1 |
1776 |
0.34 |
| chr3_56949638_56949992 | 0.18 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
684 |
0.75 |
| chr13_99953854_99954138 | 0.18 |
GPR183 |
G protein-coupled receptor 183 |
5663 |
0.22 |
| chr9_18296716_18296867 | 0.17 |
ADAMTSL1 |
ADAMTS-like 1 |
177101 |
0.03 |
| chr10_51500864_51501147 | 0.17 |
AGAP7 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 7 |
14678 |
0.19 |
| chr7_106822662_106822813 | 0.17 |
HBP1 |
HMG-box transcription factor 1 |
2374 |
0.31 |
| chr1_57041430_57041821 | 0.17 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
3616 |
0.34 |
| chr2_183289734_183289997 | 0.17 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
1884 |
0.49 |
| chr6_26198173_26198564 | 0.17 |
HIST1H3D |
histone cluster 1, H3d |
890 |
0.22 |
| chr1_111212431_111212582 | 0.17 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
5149 |
0.2 |
| chr9_20041214_20041376 | 0.17 |
ENSG00000266224 |
. |
211544 |
0.02 |
| chr3_170136599_170137401 | 0.17 |
CLDN11 |
claudin 11 |
145 |
0.97 |
| chr3_40518130_40518400 | 0.17 |
ZNF619 |
zinc finger protein 619 |
339 |
0.84 |
| chr11_96197527_96197689 | 0.17 |
ENSG00000200411 |
. |
10248 |
0.25 |
| chr16_1584614_1584961 | 0.17 |
TMEM204 |
transmembrane protein 204 |
1213 |
0.24 |
| chr8_90845937_90846088 | 0.17 |
ENSG00000207359 |
. |
67040 |
0.11 |
| chr15_38848503_38848654 | 0.17 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
3705 |
0.23 |
| chr15_50156073_50156224 | 0.17 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
12744 |
0.22 |
| chr8_80739322_80739697 | 0.17 |
ENSG00000238884 |
. |
27079 |
0.22 |
| chr8_94931050_94931201 | 0.17 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
13 |
0.98 |
| chr5_58883529_58883730 | 0.17 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
1304 |
0.62 |
| chrX_114924538_114924731 | 0.17 |
ENSG00000264759 |
. |
13498 |
0.18 |
| chr10_114291692_114291843 | 0.17 |
VTI1A |
vesicle transport through interaction with t-SNAREs 1A |
84751 |
0.08 |
| chr2_177021858_177022419 | 0.16 |
HOXD3 |
homeobox D3 |
3481 |
0.1 |
| chr6_127125664_127125831 | 0.16 |
ENSG00000201613 |
. |
214208 |
0.02 |
| chr4_100009284_100009464 | 0.16 |
ADH5 |
alcohol dehydrogenase 5 (class III), chi polypeptide |
528 |
0.5 |
| chr3_63338970_63339121 | 0.16 |
ENSG00000252662 |
. |
18582 |
0.26 |
| chr6_43019642_43019793 | 0.16 |
CUL7 |
cullin 7 |
1720 |
0.17 |
| chrX_28607014_28607165 | 0.16 |
IL1RAPL1 |
interleukin 1 receptor accessory protein-like 1 |
1573 |
0.57 |
| chr17_78705219_78705370 | 0.16 |
RP11-28G8.1 |
|
74138 |
0.1 |
| chr8_38323279_38323475 | 0.16 |
FGFR1 |
fibroblast growth factor receptor 1 |
804 |
0.6 |
| chr12_92532725_92532888 | 0.16 |
C12orf79 |
chromosome 12 open reading frame 79 |
2009 |
0.29 |
| chr4_175041157_175041343 | 0.16 |
FBXO8 |
F-box protein 8 |
163564 |
0.04 |
| chr3_114801119_114801270 | 0.16 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
10972 |
0.32 |
| chr14_30663660_30663811 | 0.16 |
PRKD1 |
protein kinase D1 |
2631 |
0.43 |
| chr4_135385341_135385492 | 0.16 |
PABPC4L |
poly(A) binding protein, cytoplasmic 4-like |
262513 |
0.02 |
| chr20_8841205_8841356 | 0.16 |
ENSG00000201348 |
. |
29447 |
0.23 |
| chr18_9117636_9118003 | 0.16 |
RP11-143J12.3 |
|
1944 |
0.28 |
| chr4_107773590_107773741 | 0.16 |
DKK2 |
dickkopf WNT signaling pathway inhibitor 2 |
183789 |
0.03 |
| chr13_47253658_47254162 | 0.16 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
61 |
0.99 |
| chr8_51255795_51255946 | 0.16 |
RP11-759A9.1 |
|
37705 |
0.2 |
| chr1_111212136_111212287 | 0.16 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
5444 |
0.2 |
| chr11_111794495_111794677 | 0.16 |
CRYAB |
crystallin, alpha B |
140 |
0.91 |
| chr1_205717316_205717519 | 0.16 |
NUCKS1 |
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
1987 |
0.26 |
| chr22_31317515_31317990 | 0.16 |
MORC2-AS1 |
MORC2 antisense RNA 1 |
543 |
0.73 |
| chr14_53575940_53576287 | 0.16 |
DDHD1 |
DDHD domain containing 1 |
40691 |
0.14 |
| chr11_128559988_128560195 | 0.16 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
2298 |
0.26 |
| chr7_122115813_122115964 | 0.16 |
CADPS2 |
Ca++-dependent secretion activator 2 |
37458 |
0.19 |
| chr14_50862384_50862648 | 0.16 |
CDKL1 |
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
101 |
0.97 |
| chr22_50318944_50319095 | 0.16 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
6638 |
0.17 |
| chr3_73606637_73606788 | 0.16 |
PDZRN3 |
PDZ domain containing ring finger 3 |
4057 |
0.29 |
| chr21_39807445_39807798 | 0.16 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
62724 |
0.14 |
| chr10_63999686_63999837 | 0.16 |
RTKN2 |
rhotekin 2 |
3739 |
0.34 |
| chr2_204572133_204572358 | 0.16 |
CD28 |
CD28 molecule |
829 |
0.71 |
| chr7_94025506_94025990 | 0.16 |
COL1A2 |
collagen, type I, alpha 2 |
1875 |
0.46 |
| chr14_52778615_52778779 | 0.16 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
2326 |
0.38 |
| chr10_123733699_123733904 | 0.15 |
NSMCE4A |
non-SMC element 4 homolog A (S. cerevisiae) |
472 |
0.82 |
| chr6_154691911_154692077 | 0.15 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
14068 |
0.27 |
| chr6_143501955_143502106 | 0.15 |
AIG1 |
androgen-induced 1 |
54643 |
0.14 |
| chr1_10341352_10341503 | 0.15 |
ENSG00000199562 |
. |
17597 |
0.15 |
| chr12_14520726_14520877 | 0.15 |
ATF7IP |
activating transcription factor 7 interacting protein |
494 |
0.85 |
| chr14_71933799_71933950 | 0.15 |
ENSG00000242330 |
. |
18468 |
0.23 |
| chrX_53356672_53356823 | 0.15 |
ENSG00000207408 |
. |
2980 |
0.23 |
| chr8_101731752_101732025 | 0.15 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
1770 |
0.27 |
| chr4_39032194_39032374 | 0.15 |
TMEM156 |
transmembrane protein 156 |
1757 |
0.38 |
| chr8_135790318_135790469 | 0.15 |
ENSG00000207582 |
. |
22457 |
0.21 |
| chr15_44120227_44120469 | 0.15 |
WDR76 |
WD repeat domain 76 |
1162 |
0.29 |
| chr7_18794250_18794418 | 0.15 |
ENSG00000222164 |
. |
53568 |
0.17 |
| chr2_197662576_197662727 | 0.15 |
GTF3C3 |
general transcription factor IIIC, polypeptide 3, 102kDa |
1732 |
0.37 |
| chr3_12436703_12437398 | 0.15 |
PPARG |
peroxisome proliferator-activated receptor gamma |
44079 |
0.15 |
| chr2_155000932_155001083 | 0.15 |
ENSG00000266512 |
. |
1713 |
0.51 |
| chr3_95425254_95425405 | 0.15 |
ENSG00000221477 |
. |
460417 |
0.01 |
| chr3_121964996_121965193 | 0.15 |
ENSG00000252170 |
. |
481 |
0.79 |
| chr3_135284920_135285071 | 0.15 |
ENSG00000253004 |
. |
233627 |
0.02 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:1904035 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0002913 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.2 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0072007 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.1 | GO:0002860 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:2000644 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.0 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.2 | GO:0010510 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0072203 | cell proliferation involved in metanephros development(GO:0072203) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.1 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.0 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.0 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.1 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.0 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.0 | GO:1901978 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0000080 | mitotic G1 phase(GO:0000080) |
| 0.0 | 0.1 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.0 | GO:0031579 | membrane raft organization(GO:0031579) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.0 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.0 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:1900078 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0010829 | negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.0 | GO:0048867 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.0 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.0 | GO:0014850 | response to muscle activity(GO:0014850) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.0 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |