Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for HOXD1
Z-value: 0.86
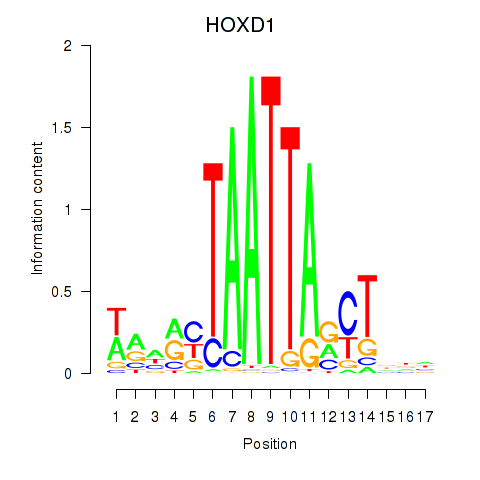
Transcription factors associated with HOXD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD1
|
ENSG00000128645.11 | homeobox D1 |
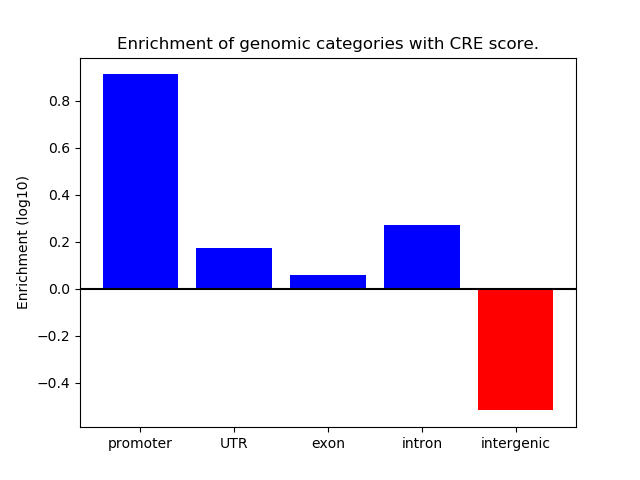
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_177053046_177053572 | HOXD1 | 2 | 0.507034 | -0.31 | 4.2e-01 | Click! |
Activity of the HOXD1 motif across conditions
Conditions sorted by the z-value of the HOXD1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
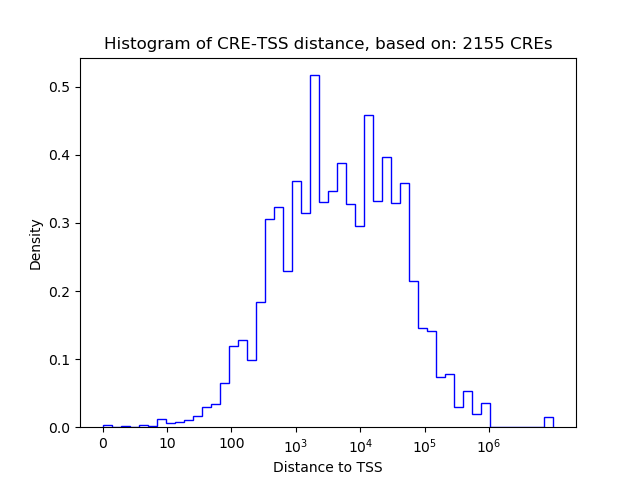
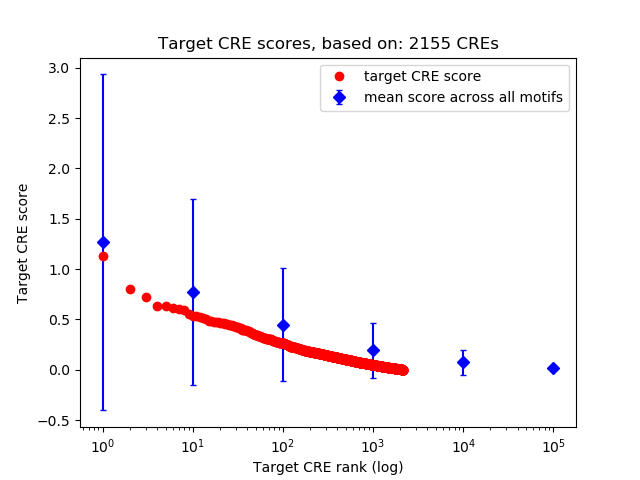
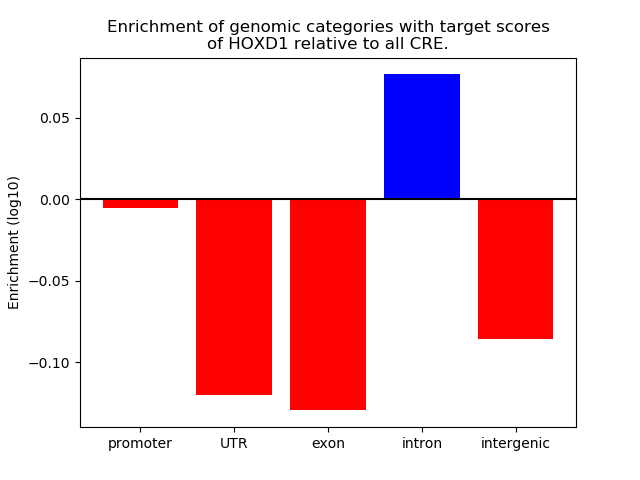
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_19815653_19816006 | 1.13 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
2040 |
0.46 |
| chr21_15917916_15918619 | 0.80 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
395 |
0.89 |
| chr2_193992778_193992929 | 0.73 |
TMEFF2 |
transmembrane protein with EGF-like and two follistatin-like domains 2 |
932418 |
0.0 |
| chr17_37933702_37934037 | 0.63 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
609 |
0.66 |
| chr14_22947593_22947744 | 0.63 |
TRAJ60 |
T cell receptor alpha joining 60 (pseudogene) |
2372 |
0.15 |
| chr13_99957828_99957979 | 0.61 |
GPR183 |
G protein-coupled receptor 183 |
1756 |
0.38 |
| chr3_186745077_186745229 | 0.60 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
1882 |
0.42 |
| chrX_28529887_28530038 | 0.59 |
IL1RAPL1 |
interleukin 1 receptor accessory protein-like 1 |
75554 |
0.12 |
| chr1_29253817_29254129 | 0.55 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
12882 |
0.18 |
| chrX_31453419_31453570 | 0.53 |
ENSG00000252903 |
. |
87185 |
0.1 |
| chr1_66806017_66806168 | 0.53 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
8220 |
0.31 |
| chr4_144312492_144312643 | 0.52 |
GAB1 |
GRB2-associated binding protein 1 |
96 |
0.98 |
| chr3_108543496_108543820 | 0.52 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
2039 |
0.42 |
| chr17_75456326_75456913 | 0.50 |
SEPT9 |
septin 9 |
4171 |
0.18 |
| chr13_40532306_40532606 | 0.49 |
ENSG00000212553 |
. |
101092 |
0.08 |
| chr21_32555155_32555306 | 0.48 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
52691 |
0.16 |
| chrX_78400540_78401325 | 0.47 |
GPR174 |
G protein-coupled receptor 174 |
25537 |
0.27 |
| chr4_25865391_25865542 | 0.47 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
84 |
0.98 |
| chr5_126094279_126094440 | 0.47 |
ENSG00000252185 |
. |
3250 |
0.29 |
| chr7_106822662_106822813 | 0.47 |
HBP1 |
HMG-box transcription factor 1 |
2374 |
0.31 |
| chr15_60874237_60874501 | 0.46 |
RORA |
RAR-related orphan receptor A |
10371 |
0.22 |
| chr2_235359483_235359634 | 0.46 |
ARL4C |
ADP-ribosylation factor-like 4C |
45686 |
0.21 |
| chr4_143321296_143321548 | 0.46 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
30990 |
0.26 |
| chr9_80524074_80524374 | 0.45 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
86309 |
0.1 |
| chr3_135909805_135909956 | 0.45 |
MSL2 |
male-specific lethal 2 homolog (Drosophila) |
3516 |
0.32 |
| chr2_191274024_191274175 | 0.45 |
MFSD6 |
major facilitator superfamily domain containing 6 |
1018 |
0.56 |
| chr14_22993528_22993679 | 0.44 |
TRAJ15 |
T cell receptor alpha joining 15 |
4977 |
0.12 |
| chr14_70111468_70111619 | 0.44 |
KIAA0247 |
KIAA0247 |
33230 |
0.18 |
| chr5_118653769_118653920 | 0.43 |
ENSG00000243333 |
. |
11518 |
0.19 |
| chr1_225610206_225610357 | 0.43 |
LBR |
lamin B receptor |
5318 |
0.2 |
| chr7_26141262_26141558 | 0.42 |
ENSG00000266430 |
. |
41451 |
0.15 |
| chr10_17701952_17702103 | 0.42 |
STAM |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
15763 |
0.14 |
| chr12_47600587_47601314 | 0.42 |
PCED1B |
PC-esterase domain containing 1B |
9102 |
0.22 |
| chr13_41555714_41555943 | 0.41 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
590 |
0.77 |
| chr1_234544831_234544982 | 0.40 |
COA6 |
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
35459 |
0.12 |
| chr11_58777091_58777242 | 0.39 |
RP11-142C4.6 |
|
30408 |
0.16 |
| chr1_192546230_192546423 | 0.39 |
RGS1 |
regulator of G-protein signaling 1 |
1423 |
0.45 |
| chr12_1434091_1434389 | 0.39 |
RP5-951N9.2 |
|
60759 |
0.12 |
| chr5_35863316_35863652 | 0.39 |
IL7R |
interleukin 7 receptor |
6490 |
0.21 |
| chr6_130391698_130391967 | 0.39 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
50130 |
0.15 |
| chr4_39032194_39032374 | 0.39 |
TMEM156 |
transmembrane protein 156 |
1757 |
0.38 |
| chr1_186291714_186291865 | 0.38 |
ENSG00000202025 |
. |
10729 |
0.17 |
| chr4_99565395_99565546 | 0.38 |
TSPAN5 |
tetraspanin 5 |
13316 |
0.22 |
| chr8_27131821_27132033 | 0.36 |
STMN4 |
stathmin-like 4 |
15990 |
0.18 |
| chr6_154568354_154568864 | 0.36 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
53 |
0.99 |
| chr3_40518130_40518400 | 0.36 |
ZNF619 |
zinc finger protein 619 |
339 |
0.84 |
| chr21_25260128_25260279 | 0.36 |
ENSG00000199698 |
. |
455601 |
0.01 |
| chrX_88218836_88218987 | 0.36 |
CPXCR1 |
CPX chromosome region, candidate 1 |
216675 |
0.02 |
| chr15_93431332_93431756 | 0.35 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
5018 |
0.2 |
| chr1_160614457_160614608 | 0.35 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
2279 |
0.26 |
| chr12_12510418_12511218 | 0.34 |
LOH12CR1 |
loss of heterozygosity, 12, chromosomal region 1 |
466 |
0.63 |
| chr11_128894218_128894369 | 0.34 |
ARHGAP32 |
Rho GTPase activating protein 32 |
205 |
0.94 |
| chr4_36243817_36244102 | 0.34 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
1602 |
0.35 |
| chr15_96817385_96817602 | 0.34 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
1732 |
0.37 |
| chr7_106507057_106507208 | 0.34 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
1208 |
0.59 |
| chr5_61577589_61577740 | 0.33 |
KIF2A |
kinesin heavy chain member 2A |
24325 |
0.24 |
| chr6_106972751_106973424 | 0.33 |
AIM1 |
absent in melanoma 1 |
13357 |
0.2 |
| chr15_50648671_50648965 | 0.32 |
ENSG00000244879 |
. |
150 |
0.91 |
| chr2_238893215_238893433 | 0.32 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
11587 |
0.2 |
| chr13_21050204_21050392 | 0.32 |
ENSG00000263978 |
. |
42313 |
0.15 |
| chr6_41855450_41855601 | 0.31 |
USP49 |
ubiquitin specific peptidase 49 |
458 |
0.75 |
| chr3_95425254_95425405 | 0.31 |
ENSG00000221477 |
. |
460417 |
0.01 |
| chr7_156761668_156761819 | 0.31 |
NOM1 |
nucleolar protein with MIF4G domain 1 |
19326 |
0.16 |
| chr4_154411300_154411567 | 0.31 |
KIAA0922 |
KIAA0922 |
23932 |
0.22 |
| chr14_34521580_34521731 | 0.31 |
EGLN3-AS1 |
EGLN3 antisense RNA 1 |
12352 |
0.27 |
| chr7_37011253_37011404 | 0.31 |
ELMO1 |
engulfment and cell motility 1 |
13337 |
0.2 |
| chr6_11727810_11727987 | 0.31 |
ADTRP |
androgen-dependent TFPI-regulating protein |
7991 |
0.25 |
| chr21_45341679_45341830 | 0.31 |
ENSG00000199598 |
. |
1972 |
0.29 |
| chr7_17039488_17039788 | 0.30 |
AGR3 |
anterior gradient 3 |
118027 |
0.06 |
| chr13_45991538_45991858 | 0.30 |
SLC25A30 |
solute carrier family 25, member 30 |
811 |
0.6 |
| chr2_68379959_68380110 | 0.30 |
WDR92 |
WD repeat domain 92 |
4571 |
0.18 |
| chr2_213997408_213997559 | 0.30 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
15870 |
0.28 |
| chrX_78403286_78403508 | 0.30 |
GPR174 |
G protein-coupled receptor 174 |
23072 |
0.28 |
| chr4_140587624_140588180 | 0.30 |
MGST2 |
microsomal glutathione S-transferase 2 |
913 |
0.61 |
| chr1_158983210_158983361 | 0.30 |
IFI16 |
interferon, gamma-inducible protein 16 |
1186 |
0.5 |
| chr12_46062870_46063096 | 0.29 |
ARID2 |
AT rich interactive domain 2 (ARID, RFX-like) |
60465 |
0.15 |
| chrY_1551045_1551310 | 0.29 |
NA |
NA |
> 106 |
NA |
| chr10_129861850_129862130 | 0.29 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
16156 |
0.26 |
| chr4_105887659_105887892 | 0.29 |
ENSG00000251906 |
. |
8044 |
0.31 |
| chr3_156400615_156400766 | 0.28 |
TIPARP |
TCDD-inducible poly(ADP-ribose) polymerase |
1735 |
0.41 |
| chr13_97879341_97880219 | 0.28 |
MBNL2 |
muscleblind-like splicing regulator 2 |
5171 |
0.33 |
| chr12_14924776_14925101 | 0.28 |
HIST4H4 |
histone cluster 4, H4 |
873 |
0.43 |
| chr10_90752530_90752985 | 0.28 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
1610 |
0.28 |
| chr21_36367168_36367319 | 0.28 |
RUNX1 |
runt-related transcription factor 1 |
54219 |
0.18 |
| chr1_206646546_206646697 | 0.28 |
IKBKE |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
2783 |
0.21 |
| chr16_71935673_71935824 | 0.28 |
IST1 |
increased sodium tolerance 1 homolog (yeast) |
6256 |
0.13 |
| chr10_70850849_70851349 | 0.28 |
SRGN |
serglycin |
3225 |
0.25 |
| chr11_20388995_20389167 | 0.27 |
HTATIP2 |
HIV-1 Tat interactive protein 2, 30kDa |
3355 |
0.31 |
| chr9_24621401_24621552 | 0.27 |
IZUMO3 |
IZUMO family member 3 |
75532 |
0.13 |
| chr18_2605370_2605521 | 0.27 |
NDC80 |
NDC80 kinetochore complex component |
5335 |
0.16 |
| chr5_68338922_68339492 | 0.27 |
SLC30A5 |
solute carrier family 30 (zinc transporter), member 5 |
50266 |
0.12 |
| chr3_108549304_108549455 | 0.27 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
7760 |
0.27 |
| chr18_60982497_60982692 | 0.27 |
RP11-28F1.2 |
|
1279 |
0.41 |
| chr11_88068800_88069370 | 0.27 |
CTSC |
cathepsin C |
1816 |
0.49 |
| chr8_103807257_103807446 | 0.27 |
ENSG00000266799 |
. |
61404 |
0.1 |
| chrX_117750069_117750220 | 0.27 |
ENSG00000206862 |
. |
46666 |
0.16 |
| chr4_36245078_36245884 | 0.27 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
80 |
0.74 |
| chr1_246488003_246488154 | 0.26 |
SMYD3-IT1 |
SMYD3 intronic transcript 1 (non-protein coding) |
2338 |
0.35 |
| chrX_23969833_23969984 | 0.26 |
CXorf58 |
chromosome X open reading frame 58 |
41408 |
0.14 |
| chr6_98593876_98594027 | 0.26 |
ENSG00000238367 |
. |
121544 |
0.07 |
| chr3_171848594_171848745 | 0.26 |
FNDC3B |
fibronectin type III domain containing 3B |
3905 |
0.33 |
| chr10_17548651_17548924 | 0.26 |
ST8SIA6 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
52458 |
0.13 |
| chr11_128589269_128589524 | 0.26 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
23478 |
0.17 |
| chrX_78620075_78620267 | 0.26 |
ITM2A |
integral membrane protein 2A |
2685 |
0.44 |
| chrX_78200973_78201449 | 0.26 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
293 |
0.95 |
| chr11_118900356_118900707 | 0.26 |
SLC37A4 |
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
1040 |
0.25 |
| chr8_66742389_66742540 | 0.25 |
PDE7A |
phosphodiesterase 7A |
8519 |
0.29 |
| chr14_22461559_22461710 | 0.25 |
ENSG00000238634 |
. |
149253 |
0.04 |
| chr19_48832910_48833240 | 0.25 |
EMP3 |
epithelial membrane protein 3 |
4210 |
0.13 |
| chr12_131796680_131796831 | 0.25 |
ENSG00000212251 |
. |
12459 |
0.23 |
| chr7_91705589_91705740 | 0.25 |
AKAP9 |
A kinase (PRKA) anchor protein 9 |
6212 |
0.22 |
| chr4_166361643_166362016 | 0.25 |
CPE |
carboxypeptidase E |
22451 |
0.23 |
| chr15_52844361_52844512 | 0.25 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
5505 |
0.2 |
| chr6_91004568_91004758 | 0.24 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
1798 |
0.43 |
| chr14_90084669_90085459 | 0.24 |
FOXN3 |
forkhead box N3 |
410 |
0.78 |
| chr20_25023152_25023303 | 0.24 |
ACSS1 |
acyl-CoA synthetase short-chain family member 1 |
9888 |
0.2 |
| chr11_119572371_119572522 | 0.23 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
26817 |
0.16 |
| chr9_125662893_125663901 | 0.23 |
RC3H2 |
ring finger and CCCH-type domains 2 |
3008 |
0.16 |
| chr12_94805998_94806149 | 0.23 |
CCDC41 |
coiled-coil domain containing 41 |
47649 |
0.14 |
| chr3_187492063_187492306 | 0.23 |
BCL6 |
B-cell CLL/lymphoma 6 |
28669 |
0.2 |
| chr12_54679325_54679476 | 0.23 |
HNRNPA1 |
heterogeneous nuclear ribonucleoprotein A1 |
3418 |
0.1 |
| chr6_74231834_74232090 | 0.23 |
EEF1A1 |
eukaryotic translation elongation factor 1 alpha 1 |
339 |
0.77 |
| chr1_247553346_247553582 | 0.23 |
NLRP3 |
NLR family, pyrin domain containing 3 |
25994 |
0.15 |
| chr6_17972440_17972591 | 0.23 |
KIF13A |
kinesin family member 13A |
15179 |
0.27 |
| chr5_116293290_116293441 | 0.23 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
382735 |
0.01 |
| chr12_64798382_64799443 | 0.23 |
XPOT |
exportin, tRNA |
86 |
0.97 |
| chr20_48328141_48328292 | 0.23 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
2199 |
0.3 |
| chr11_20179312_20179463 | 0.23 |
DBX1 |
developing brain homeobox 1 |
2483 |
0.35 |
| chr14_71454929_71455080 | 0.23 |
PCNX |
pecanex homolog (Drosophila) |
24744 |
0.26 |
| chr17_54857369_54857761 | 0.23 |
C17orf67 |
chromosome 17 open reading frame 67 |
35685 |
0.15 |
| chr14_88471433_88472132 | 0.23 |
GPR65 |
G protein-coupled receptor 65 |
314 |
0.88 |
| chr17_47766049_47766200 | 0.23 |
SPOP |
speckle-type POZ protein |
10528 |
0.14 |
| chr12_64852912_64853865 | 0.22 |
TBK1 |
TANK-binding kinase 1 |
7133 |
0.2 |
| chr21_16726840_16726991 | 0.22 |
ENSG00000212564 |
. |
259687 |
0.02 |
| chr10_119202905_119203056 | 0.22 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
33704 |
0.18 |
| chr5_75600050_75600201 | 0.22 |
RP11-466P24.6 |
|
7162 |
0.3 |
| chr3_71539044_71539195 | 0.22 |
ENSG00000221264 |
. |
52121 |
0.14 |
| chr12_96794557_96795184 | 0.22 |
CDK17 |
cyclin-dependent kinase 17 |
532 |
0.79 |
| chr1_226919798_226919959 | 0.22 |
ITPKB |
inositol-trisphosphate 3-kinase B |
5281 |
0.26 |
| chr8_42034876_42035027 | 0.22 |
AP3M2 |
adaptor-related protein complex 3, mu 2 subunit |
10144 |
0.16 |
| chr20_3452818_3452969 | 0.22 |
ATRN |
attractin |
1138 |
0.49 |
| chr11_88087417_88087693 | 0.22 |
CTSC |
cathepsin C |
16600 |
0.28 |
| chr2_161995560_161995711 | 0.22 |
TANK |
TRAF family member-associated NFKB activator |
2169 |
0.37 |
| chr6_90066974_90067553 | 0.22 |
UBE2J1 |
ubiquitin-conjugating enzyme E2, J1 |
4696 |
0.23 |
| chr6_143179346_143179497 | 0.21 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
21237 |
0.25 |
| chr18_9404580_9404814 | 0.21 |
TWSG1 |
twisted gastrulation BMP signaling modulator 1 |
69849 |
0.08 |
| chrX_1600809_1601304 | 0.21 |
ASMTL |
acetylserotonin O-methyltransferase-like |
28401 |
0.16 |
| chrX_18516169_18516320 | 0.21 |
CDKL5 |
cyclin-dependent kinase-like 5 |
55936 |
0.14 |
| chr15_29021803_29021954 | 0.21 |
RP11-578F21.10 |
|
11870 |
0.15 |
| chr5_110560941_110561277 | 0.21 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
1325 |
0.51 |
| chr10_63753889_63754171 | 0.21 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
54940 |
0.15 |
| chr6_26198173_26198564 | 0.21 |
HIST1H3D |
histone cluster 1, H3d |
890 |
0.22 |
| chr10_106007564_106007715 | 0.21 |
GSTO1 |
glutathione S-transferase omega 1 |
6313 |
0.13 |
| chr1_29254771_29254973 | 0.21 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
13781 |
0.18 |
| chr7_104658006_104658503 | 0.20 |
KMT2E |
lysine (K)-specific methyltransferase 2E |
3346 |
0.19 |
| chr4_100864856_100865007 | 0.20 |
DNAJB14 |
DnaJ (Hsp40) homolog, subfamily B, member 14 |
2952 |
0.23 |
| chr4_153268296_153268447 | 0.20 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
1587 |
0.42 |
| chr1_11335043_11335194 | 0.20 |
UBIAD1 |
UbiA prenyltransferase domain containing 1 |
1127 |
0.47 |
| chr3_45732221_45732372 | 0.20 |
SACM1L |
SAC1 suppressor of actin mutations 1-like (yeast) |
1345 |
0.36 |
| chr1_10516682_10516833 | 0.20 |
RP5-1113E3.3 |
|
1855 |
0.2 |
| chr15_52023462_52024365 | 0.20 |
LYSMD2 |
LysM, putative peptidoglycan-binding, domain containing 2 |
6405 |
0.15 |
| chr10_70230757_70231440 | 0.20 |
DNA2 |
DNA replication helicase/nuclease 2 |
545 |
0.71 |
| chr5_74871506_74871657 | 0.20 |
CTC-366B18.2 |
|
23624 |
0.14 |
| chr2_113945705_113945856 | 0.20 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
8077 |
0.14 |
| chr16_53133173_53134292 | 0.20 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
660 |
0.77 |
| chrX_83325612_83325763 | 0.20 |
RPS6KA6 |
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
117228 |
0.07 |
| chr13_46755198_46755455 | 0.20 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
1133 |
0.45 |
| chr2_37576303_37576454 | 0.19 |
QPCT |
glutaminyl-peptide cyclotransferase |
4526 |
0.2 |
| chr22_17956680_17956831 | 0.19 |
CECR2 |
cat eye syndrome chromosome region, candidate 2 |
128 |
0.97 |
| chr2_38822967_38823118 | 0.19 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
4126 |
0.22 |
| chr8_27223967_27224118 | 0.19 |
PTK2B |
protein tyrosine kinase 2 beta |
14126 |
0.22 |
| chr17_75178097_75178354 | 0.19 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
1106 |
0.55 |
| chr7_115668295_115668446 | 0.19 |
TFEC |
transcription factor EC |
2425 |
0.43 |
| chr3_151961363_151961565 | 0.19 |
MBNL1 |
muscleblind-like splicing regulator 1 |
24365 |
0.2 |
| chr9_16776546_16776697 | 0.19 |
BNC2 |
basonuclin 2 |
48409 |
0.16 |
| chr10_3514525_3515428 | 0.19 |
RP11-184A2.3 |
|
278283 |
0.01 |
| chr1_52344711_52345319 | 0.19 |
NRD1 |
nardilysin (N-arginine dibasic convertase) |
538 |
0.71 |
| chr1_90059823_90059974 | 0.19 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
26098 |
0.15 |
| chr16_79652209_79652360 | 0.19 |
MAF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
17673 |
0.26 |
| chr17_76438588_76438739 | 0.19 |
AC061992.1 |
Uncharacterized protein |
16254 |
0.14 |
| chr10_30745504_30745655 | 0.19 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
17828 |
0.21 |
| chr1_90290694_90290845 | 0.19 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
3289 |
0.28 |
| chr1_92269176_92269391 | 0.19 |
ENSG00000239794 |
. |
26348 |
0.22 |
| chr1_150294067_150294674 | 0.19 |
PRPF3 |
pre-mRNA processing factor 3 |
368 |
0.78 |
| chr8_26184784_26184935 | 0.19 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
33860 |
0.18 |
| chr12_92534627_92535592 | 0.19 |
C12orf79 |
chromosome 12 open reading frame 79 |
1335 |
0.39 |
| chr8_102786541_102786692 | 0.19 |
NCALD |
neurocalcin delta |
14610 |
0.25 |
| chr18_74837172_74837323 | 0.19 |
MBP |
myelin basic protein |
2421 |
0.42 |
| chr1_111420872_111421052 | 0.19 |
CD53 |
CD53 molecule |
5186 |
0.22 |
| chr2_64369947_64371114 | 0.19 |
AC074289.1 |
|
157 |
0.93 |
| chr15_43531850_43532099 | 0.18 |
ENSG00000202211 |
. |
7904 |
0.13 |
| chr14_32672268_32672419 | 0.18 |
ENSG00000202337 |
. |
132 |
0.94 |
| chr14_70102783_70102934 | 0.18 |
KIAA0247 |
KIAA0247 |
24545 |
0.21 |
| chr5_81762775_81762926 | 0.18 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
161684 |
0.04 |
| chr2_106473228_106473435 | 0.18 |
AC009505.2 |
|
129 |
0.97 |
| chr13_28500606_28500757 | 0.18 |
PDX1-AS1 |
PDX1 antisense RNA 1 |
5140 |
0.16 |
| chr4_40202041_40202542 | 0.18 |
RHOH |
ras homolog family member H |
327 |
0.9 |
| chr4_15146065_15146216 | 0.18 |
ENSG00000199420 |
. |
50422 |
0.16 |
| chr2_103506378_103506656 | 0.18 |
TMEM182 |
transmembrane protein 182 |
128045 |
0.06 |
| chr5_145562456_145562664 | 0.18 |
LARS |
leucyl-tRNA synthetase |
337 |
0.89 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.2 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:1904019 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.1 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0061117 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.2 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.6 | GO:1901185 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0002423 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.2 | GO:0030260 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.1 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.0 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.0 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0007141 | male meiosis I(GO:0007141) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0031211 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.5 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 1.0 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.2 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.0 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.0 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.0 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |