Project
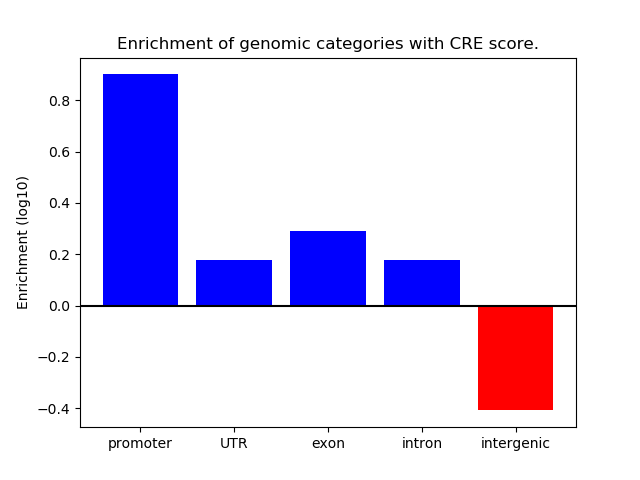
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
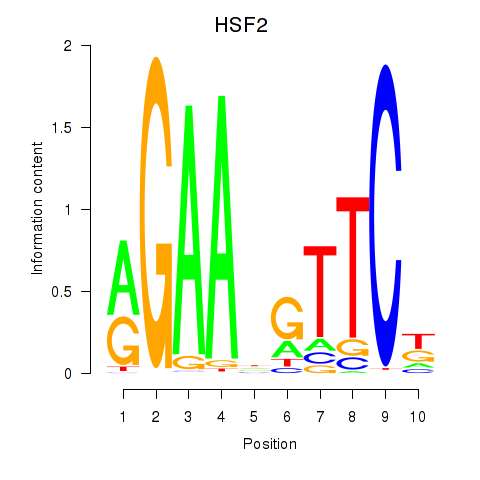
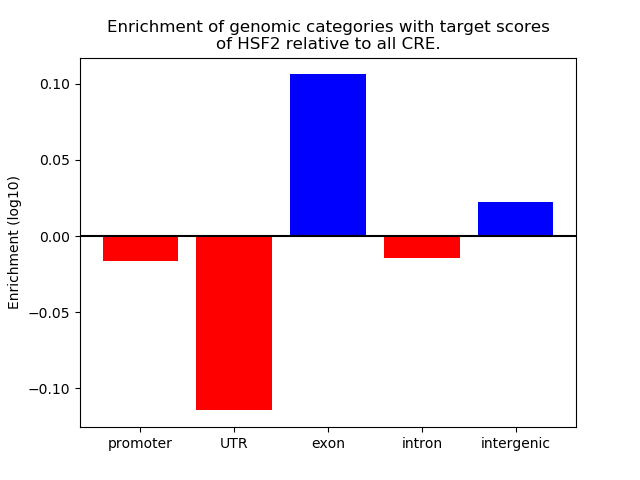
Results for HSF2
Z-value: 0.34
Transcription factors associated with HSF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF2
|
ENSG00000025156.8 | heat shock transcription factor 2 |
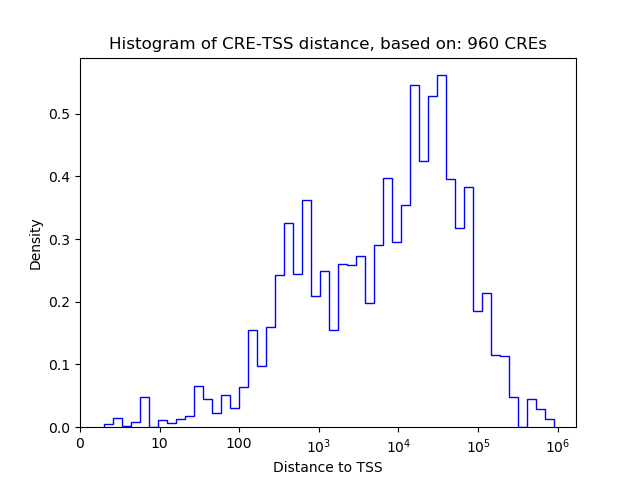
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_122720872_122721541 | HSF2 | 510 | 0.857717 | -0.80 | 9.7e-03 | Click! |
| chr6_122720489_122720640 | HSF2 | 127 | 0.976720 | -0.75 | 1.9e-02 | Click! |
| chr6_122729994_122730284 | HSF2 | 9443 | 0.258507 | -0.73 | 2.6e-02 | Click! |
| chr6_122697833_122697984 | HSF2 | 22783 | 0.244110 | 0.63 | 7.2e-02 | Click! |
| chr6_122721780_122721942 | HSF2 | 1165 | 0.602362 | 0.54 | 1.4e-01 | Click! |
Activity of the HSF2 motif across conditions
Conditions sorted by the z-value of the HSF2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
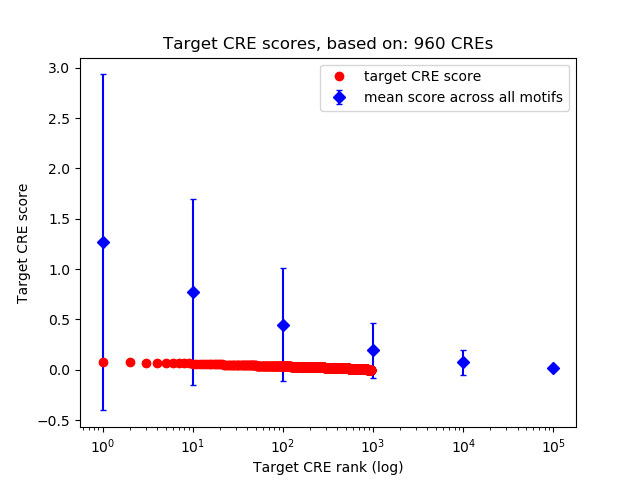
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_138866633_138866868 | 0.08 |
NHSL1 |
NHS-like 1 |
98 |
0.98 |
| chr11_117073075_117073344 | 0.07 |
TAGLN |
transgelin |
620 |
0.64 |
| chr3_49237110_49237583 | 0.07 |
CCDC36 |
coiled-coil domain containing 36 |
403 |
0.74 |
| chr17_79317268_79317735 | 0.07 |
TMEM105 |
transmembrane protein 105 |
13027 |
0.13 |
| chr5_119959781_119959971 | 0.07 |
PRR16 |
proline rich 16 |
7093 |
0.32 |
| chr19_49590923_49591074 | 0.07 |
SNRNP70 |
small nuclear ribonucleoprotein 70kDa (U1) |
1261 |
0.2 |
| chr7_120630066_120630217 | 0.07 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
465 |
0.82 |
| chr8_123801020_123801334 | 0.07 |
ZHX2 |
zinc fingers and homeoboxes 2 |
7544 |
0.22 |
| chr19_36247573_36247956 | 0.06 |
HSPB6 |
heat shock protein, alpha-crystallin-related, B6 |
146 |
0.84 |
| chr2_240966839_240966990 | 0.06 |
NDUFA10 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa |
2095 |
0.24 |
| chr9_130855866_130856180 | 0.06 |
RP11-379C10.4 |
|
713 |
0.45 |
| chr12_109123612_109123767 | 0.06 |
CORO1C |
coronin, actin binding protein, 1C |
638 |
0.67 |
| chr4_22517802_22518220 | 0.06 |
GPR125 |
G protein-coupled receptor 125 |
334 |
0.94 |
| chr17_59222769_59222920 | 0.06 |
RP11-136H19.1 |
|
9519 |
0.23 |
| chr8_49994861_49995012 | 0.06 |
C8orf22 |
chromosome 8 open reading frame 22 |
10023 |
0.32 |
| chrX_47445267_47445418 | 0.05 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
601 |
0.58 |
| chrX_10727306_10727511 | 0.05 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
81629 |
0.11 |
| chr2_175662837_175662988 | 0.05 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
33712 |
0.18 |
| chr2_85833211_85833461 | 0.05 |
TMEM150A |
transmembrane protein 150A |
3515 |
0.11 |
| chr11_48085341_48085729 | 0.05 |
ENSG00000263693 |
. |
32799 |
0.16 |
| chrX_114827823_114827974 | 0.05 |
PLS3 |
plastin 3 |
33 |
0.98 |
| chr21_27762391_27762594 | 0.05 |
AP001596.6 |
|
37 |
0.98 |
| chr14_104123773_104123924 | 0.05 |
KLC1 |
kinesin light chain 1 |
15844 |
0.09 |
| chr8_77317569_77317720 | 0.05 |
ENSG00000222231 |
. |
139677 |
0.05 |
| chr10_128352295_128352662 | 0.05 |
C10orf90 |
chromosome 10 open reading frame 90 |
6537 |
0.3 |
| chr9_114061378_114061529 | 0.05 |
OR2K2 |
olfactory receptor, family 2, subfamily K, member 2 |
29260 |
0.21 |
| chr8_6539493_6539644 | 0.05 |
CTD-2541M15.1 |
|
26132 |
0.18 |
| chr9_120467136_120467336 | 0.05 |
TLR4 |
toll-like receptor 4 |
586 |
0.75 |
| chr1_32015163_32015427 | 0.05 |
RP11-73M7.1 |
|
25879 |
0.13 |
| chr3_70121343_70121525 | 0.05 |
RP11-231I13.2 |
|
134508 |
0.05 |
| chr16_88220519_88220720 | 0.05 |
BANP |
BTG3 associated nuclear protein |
216995 |
0.02 |
| chr3_186539606_186539934 | 0.05 |
RP11-573D15.3 |
|
14289 |
0.08 |
| chr6_108020034_108020185 | 0.05 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
33490 |
0.22 |
| chrX_48768206_48768869 | 0.05 |
SLC35A2 |
solute carrier family 35 (UDP-galactose transporter), member A2 |
376 |
0.74 |
| chr18_12421527_12421678 | 0.05 |
SLMO1 |
slowmo homolog 1 (Drosophila) |
317 |
0.88 |
| chr6_117852473_117852851 | 0.05 |
DCBLD1 |
discoidin, CUB and LCCL domain containing 1 |
48837 |
0.12 |
| chr13_96704851_96705002 | 0.05 |
UGGT2 |
UDP-glucose glycoprotein glucosyltransferase 2 |
711 |
0.77 |
| chr6_136359997_136360148 | 0.05 |
RP13-143G15.3 |
|
25704 |
0.21 |
| chr4_187025490_187026623 | 0.05 |
FAM149A |
family with sequence similarity 149, member A |
175 |
0.94 |
| chr2_163099011_163099583 | 0.05 |
FAP |
fibroblast activation protein, alpha |
261 |
0.94 |
| chr3_183167571_183167804 | 0.05 |
LINC00888 |
long intergenic non-protein coding RNA 888 |
1842 |
0.19 |
| chr3_99375722_99375873 | 0.04 |
COL8A1 |
collagen, type VIII, alpha 1 |
18343 |
0.23 |
| chr5_141700925_141701122 | 0.04 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
2724 |
0.3 |
| chr19_46110497_46110665 | 0.04 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
5111 |
0.1 |
| chr2_192500067_192500218 | 0.04 |
NABP1 |
nucleic acid binding protein 1 |
42720 |
0.2 |
| chr10_9271510_9271661 | 0.04 |
ENSG00000212505 |
. |
572791 |
0.0 |
| chr22_19165246_19166141 | 0.04 |
SLC25A1 |
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
341 |
0.82 |
| chr15_30920784_30920977 | 0.04 |
ARHGAP11B |
Rho GTPase activating protein 11B |
2001 |
0.15 |
| chr1_240501387_240501653 | 0.04 |
ENSG00000252317 |
. |
3165 |
0.32 |
| chr16_74204020_74204218 | 0.04 |
PSMD7 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
126554 |
0.05 |
| chr8_68304896_68305241 | 0.04 |
ARFGEF1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
49156 |
0.18 |
| chr18_32172945_32173175 | 0.04 |
DTNA |
dystrobrevin, alpha |
222 |
0.97 |
| chr17_64187141_64188685 | 0.04 |
CEP112 |
centrosomal protein 112kDa |
60 |
0.98 |
| chr4_81207576_81207727 | 0.04 |
FGF5 |
fibroblast growth factor 5 |
19858 |
0.22 |
| chr7_128171824_128173030 | 0.04 |
RP11-274B21.1 |
|
1279 |
0.43 |
| chr1_184857651_184857802 | 0.04 |
ENSG00000252222 |
. |
67103 |
0.11 |
| chr1_158770334_158770485 | 0.04 |
OR6N2 |
olfactory receptor, family 6, subfamily N, member 2 |
22984 |
0.14 |
| chr18_10058041_10058330 | 0.04 |
ENSG00000263630 |
. |
52991 |
0.15 |
| chrX_99904753_99905093 | 0.04 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
5708 |
0.19 |
| chr18_22869258_22869420 | 0.04 |
CTD-2006O16.2 |
|
12751 |
0.28 |
| chr21_17489674_17489828 | 0.04 |
ENSG00000252273 |
. |
81922 |
0.11 |
| chr3_149258751_149258902 | 0.04 |
ENSG00000252581 |
. |
15408 |
0.19 |
| chr8_18712658_18712809 | 0.04 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
849 |
0.7 |
| chr9_124888898_124889336 | 0.04 |
ENSG00000266583 |
. |
6671 |
0.17 |
| chr8_15095473_15095828 | 0.04 |
SGCZ |
sarcoglycan, zeta |
198 |
0.97 |
| chr4_139935567_139935733 | 0.04 |
CCRN4L |
CCR4 carbon catabolite repression 4-like (S. cerevisiae) |
1293 |
0.33 |
| chr8_48103892_48104098 | 0.04 |
IGLV8OR8-1 |
immunoglobulin lambda variable 8/OR8-1 (pseudogene) |
10525 |
0.24 |
| chr17_13503336_13503489 | 0.04 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
1832 |
0.45 |
| chr8_120650260_120650433 | 0.04 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
674 |
0.78 |
| chr10_4814227_4814378 | 0.04 |
AKR1E2 |
aldo-keto reductase family 1, member E2 |
14519 |
0.27 |
| chr8_93113958_93114847 | 0.04 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
1052 |
0.69 |
| chr6_22025873_22026078 | 0.04 |
ENSG00000222515 |
. |
59798 |
0.16 |
| chr1_243906024_243906249 | 0.04 |
RP11-370K11.1 |
|
2013 |
0.42 |
| chr2_85936608_85936759 | 0.04 |
GNLY |
granulysin |
14192 |
0.13 |
| chr19_28500299_28500450 | 0.04 |
AC022153.1 |
|
72705 |
0.13 |
| chr20_44750642_44750793 | 0.04 |
CD40 |
CD40 molecule, TNF receptor superfamily member 5 |
3762 |
0.23 |
| chrX_102940107_102940387 | 0.04 |
MORF4L2 |
mortality factor 4 like 2 |
902 |
0.43 |
| chr7_27848119_27848315 | 0.04 |
TAX1BP1 |
Tax1 (human T-cell leukemia virus type I) binding protein 1 |
15460 |
0.23 |
| chr8_68418566_68418743 | 0.04 |
ARFGEF1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
162742 |
0.04 |
| chr4_175750179_175750687 | 0.04 |
GLRA3 |
glycine receptor, alpha 3 |
32 |
0.99 |
| chr7_22589223_22589977 | 0.04 |
AC002480.3 |
|
13356 |
0.2 |
| chr1_156829720_156830506 | 0.04 |
NTRK1 |
neurotrophic tyrosine kinase, receptor, type 1 |
494 |
0.64 |
| chr17_65256580_65256837 | 0.04 |
ENSG00000200619 |
. |
7518 |
0.14 |
| chr9_126164158_126164309 | 0.04 |
ENSG00000207991 |
. |
649 |
0.76 |
| chr6_51848179_51848330 | 0.04 |
ENSG00000240641 |
. |
7796 |
0.27 |
| chr1_23809780_23810676 | 0.04 |
ASAP3 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
443 |
0.82 |
| chr2_3223937_3224088 | 0.04 |
TSSC1-IT1 |
TSSC1 intronic transcript 1 (non-protein coding) |
81224 |
0.1 |
| chr6_90142956_90143739 | 0.04 |
ANKRD6 |
ankyrin repeat domain 6 |
415 |
0.85 |
| chr2_169733635_169733786 | 0.04 |
SPC25 |
SPC25, NDC80 kinetochore complex component |
13245 |
0.2 |
| chrX_152989287_152989823 | 0.04 |
BCAP31 |
B-cell receptor-associated protein 31 |
6 |
0.92 |
| chr11_68426343_68426647 | 0.04 |
GAL |
galanin/GMAP prepropeptide |
25448 |
0.16 |
| chr13_107572015_107572438 | 0.04 |
ENSG00000239050 |
. |
55148 |
0.17 |
| chr10_65558718_65559094 | 0.04 |
REEP3 |
receptor accessory protein 3 |
277783 |
0.01 |
| chr19_17345728_17346858 | 0.04 |
OCEL1 |
occludin/ELL domain containing 1 |
7228 |
0.08 |
| chr16_29232306_29232457 | 0.04 |
RP11-231C14.6 |
|
91290 |
0.06 |
| chr4_146403033_146403938 | 0.04 |
SMAD1 |
SMAD family member 1 |
153 |
0.96 |
| chr12_76153409_76153694 | 0.04 |
ENSG00000264062 |
. |
35230 |
0.2 |
| chr11_134818938_134819089 | 0.04 |
AP003062.1 |
CDNA FLJ27342 fis, clone TST02993; Uncharacterized protein |
36233 |
0.24 |
| chr19_49521308_49522757 | 0.04 |
LHB |
luteinizing hormone beta polypeptide |
1694 |
0.14 |
| chr4_48702119_48702270 | 0.03 |
FRYL |
FRY-like |
19006 |
0.24 |
| chrX_2882242_2882393 | 0.03 |
ARSE |
arylsulfatase E (chondrodysplasia punctata 1) |
6 |
0.98 |
| chr1_99646133_99646284 | 0.03 |
LPPR4 |
Lipid phosphate phosphatase-related protein type 4 |
83301 |
0.11 |
| chr12_51702265_51702437 | 0.03 |
BIN2 |
bridging integrator 2 |
15548 |
0.14 |
| chr2_16831792_16832248 | 0.03 |
FAM49A |
family with sequence similarity 49, member A |
15076 |
0.3 |
| chr10_44879097_44879375 | 0.03 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
1255 |
0.52 |
| chr5_178367547_178367761 | 0.03 |
ZNF454 |
zinc finger protein 454 |
538 |
0.73 |
| chr17_66050514_66050792 | 0.03 |
KPNA2 |
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
17543 |
0.17 |
| chr11_2972850_2973001 | 0.03 |
ENSG00000207008 |
. |
12198 |
0.12 |
| chr11_35627316_35627467 | 0.03 |
FJX1 |
four jointed box 1 (Drosophila) |
12344 |
0.23 |
| chr6_158490615_158490766 | 0.03 |
SYNJ2 |
synaptojanin 2 |
8022 |
0.22 |
| chr7_90896542_90897161 | 0.03 |
FZD1 |
frizzled family receptor 1 |
3068 |
0.42 |
| chr4_163085753_163086130 | 0.03 |
FSTL5 |
follistatin-like 5 |
754 |
0.8 |
| chr10_95302144_95302315 | 0.03 |
FFAR4 |
free fatty acid receptor 4 |
24193 |
0.14 |
| chr1_155830714_155830915 | 0.03 |
SYT11 |
synaptotagmin XI |
1467 |
0.25 |
| chr3_66519543_66519694 | 0.03 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
31738 |
0.24 |
| chr6_3911647_3911836 | 0.03 |
FAM50B |
family with sequence similarity 50, member B |
62119 |
0.1 |
| chr2_239800244_239800395 | 0.03 |
TWIST2 |
twist family bHLH transcription factor 2 |
43646 |
0.15 |
| chr11_110967676_110967838 | 0.03 |
ENSG00000200168 |
. |
56825 |
0.15 |
| chr11_106913477_106913628 | 0.03 |
GUCY1A2 |
guanylate cyclase 1, soluble, alpha 2 |
24302 |
0.28 |
| chr2_109273663_109273868 | 0.03 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
2256 |
0.33 |
| chr17_43170825_43171067 | 0.03 |
PLCD3 |
phospholipase C, delta 3 |
19970 |
0.1 |
| chr4_16969555_16969706 | 0.03 |
LDB2 |
LIM domain binding 2 |
69198 |
0.14 |
| chr1_19809409_19809560 | 0.03 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
1095 |
0.47 |
| chr8_18709187_18709437 | 0.03 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
2572 |
0.36 |
| chr5_31806525_31806676 | 0.03 |
PDZD2 |
PDZ domain containing 2 |
7569 |
0.14 |
| chr9_18496529_18496680 | 0.03 |
ADAMTSL1 |
ADAMTS-like 1 |
22373 |
0.25 |
| chr11_27720314_27720465 | 0.03 |
BDNF |
brain-derived neurotrophic factor |
825 |
0.7 |
| chr5_15620667_15620944 | 0.03 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
4714 |
0.28 |
| chr17_19172840_19172991 | 0.03 |
EPN2 |
epsin 2 |
11682 |
0.12 |
| chr10_104536223_104536534 | 0.03 |
WBP1L |
WW domain binding protein 1-like |
372 |
0.83 |
| chr8_97744511_97744662 | 0.03 |
CPQ |
carboxypeptidase Q |
28616 |
0.25 |
| chr2_133425157_133425453 | 0.03 |
LYPD1 |
LY6/PLAUR domain containing 1 |
2473 |
0.33 |
| chr12_66635842_66635993 | 0.03 |
ENSG00000266539 |
. |
6515 |
0.17 |
| chr18_56084210_56084361 | 0.03 |
RP11-845C23.3 |
|
16971 |
0.16 |
| chr18_499765_500605 | 0.03 |
COLEC12 |
collectin sub-family member 12 |
537 |
0.79 |
| chr2_231692712_231693467 | 0.03 |
ITM2C |
integral membrane protein 2C |
36265 |
0.14 |
| chr15_70011457_70011608 | 0.03 |
ENSG00000238870 |
. |
11629 |
0.27 |
| chr11_65200671_65200881 | 0.03 |
ENSG00000245532 |
. |
11153 |
0.12 |
| chr3_65130539_65130690 | 0.03 |
ENSG00000264871 |
. |
88044 |
0.1 |
| chr2_192711472_192711842 | 0.03 |
SDPR |
serum deprivation response |
324 |
0.55 |
| chr12_116498850_116499001 | 0.03 |
ENSG00000200665 |
. |
21450 |
0.22 |
| chr16_69960974_69961125 | 0.03 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
2156 |
0.25 |
| chr4_169564167_169564409 | 0.03 |
RP11-610J23.1 |
|
5450 |
0.22 |
| chr2_189652663_189652814 | 0.03 |
DIRC1 |
disrupted in renal carcinoma 1 |
2093 |
0.42 |
| chr21_43537496_43537668 | 0.03 |
C21orf128 |
chromosome 21 open reading frame 128 |
8938 |
0.21 |
| chr21_46714077_46714379 | 0.03 |
ENSG00000215447 |
. |
6261 |
0.19 |
| chr11_133920657_133920816 | 0.03 |
JAM3 |
junctional adhesion molecule 3 |
18084 |
0.22 |
| chr2_110658435_110658586 | 0.03 |
LIMS3 |
LIM and senescent cell antigen-like domains 3 |
2242 |
0.42 |
| chr8_17518406_17518557 | 0.03 |
MTUS1 |
microtubule associated tumor suppressor 1 |
15357 |
0.17 |
| chr11_111607263_111607414 | 0.03 |
PPP2R1B |
protein phosphatase 2, regulatory subunit A, beta |
18424 |
0.17 |
| chr11_128339811_128339962 | 0.03 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
35403 |
0.2 |
| chr5_14448576_14448727 | 0.03 |
TRIO |
trio Rho guanine nucleotide exchange factor |
39906 |
0.21 |
| chr12_77272013_77272805 | 0.03 |
CSRP2 |
cysteine and glycine-rich protein 2 |
356 |
0.56 |
| chr10_94725160_94725374 | 0.03 |
EXOC6 |
exocyst complex component 6 |
31964 |
0.2 |
| chr8_23220998_23221149 | 0.03 |
LOXL2 |
lysyl oxidase-like 2 |
7566 |
0.18 |
| chr1_98864645_98864799 | 0.03 |
ENSG00000221777 |
. |
25781 |
0.28 |
| chr21_17681697_17681848 | 0.03 |
ENSG00000201025 |
. |
24683 |
0.27 |
| chr14_77493944_77494823 | 0.03 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
651 |
0.73 |
| chr4_88419703_88419884 | 0.03 |
SPARCL1 |
SPARC-like 1 (hevin) |
29978 |
0.17 |
| chr20_48671365_48671516 | 0.03 |
UBE2V1 |
ubiquitin-conjugating enzyme E2 variant 1 |
58236 |
0.09 |
| chr1_27098835_27099154 | 0.03 |
ARID1A |
AT rich interactive domain 1A (SWI-like) |
461 |
0.73 |
| chr2_111228087_111228238 | 0.03 |
LIMS3 |
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein |
2231 |
0.25 |
| chr2_133174260_133174576 | 0.03 |
GPR39 |
G protein-coupled receptor 39 |
271 |
0.93 |
| chr17_70339032_70339432 | 0.03 |
SOX9 |
SRY (sex determining region Y)-box 9 |
222071 |
0.02 |
| chr4_184264353_184264576 | 0.03 |
ENSG00000238596 |
. |
14013 |
0.14 |
| chr13_67928310_67928532 | 0.03 |
PCDH9 |
protocadherin 9 |
123953 |
0.06 |
| chr12_63078503_63078654 | 0.03 |
ENSG00000238475 |
. |
33623 |
0.17 |
| chrX_13751428_13751579 | 0.03 |
TRAPPC2 |
trafficking protein particle complex 2 |
1172 |
0.37 |
| chr11_111413272_111413423 | 0.03 |
LAYN |
layilin |
1070 |
0.38 |
| chr14_27066556_27066724 | 0.03 |
NOVA1 |
neuro-oncological ventral antigen 1 |
2 |
0.96 |
| chr16_14109675_14109826 | 0.03 |
MKL2 |
MKL/myocardin-like 2 |
55428 |
0.14 |
| chr3_29800464_29800615 | 0.03 |
RBMS3-AS2 |
RBMS3 antisense RNA 2 |
116239 |
0.07 |
| chr8_13014143_13014294 | 0.03 |
ENSG00000200630 |
. |
3469 |
0.29 |
| chr4_159925007_159925158 | 0.03 |
ENSG00000221314 |
. |
3640 |
0.25 |
| chr1_64245163_64245314 | 0.03 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
5524 |
0.3 |
| chr3_189794073_189794333 | 0.03 |
ENSG00000265045 |
. |
37520 |
0.14 |
| chr7_87229218_87230455 | 0.03 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
336 |
0.9 |
| chr3_114168204_114168554 | 0.03 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
5151 |
0.29 |
| chr2_43188289_43188440 | 0.03 |
ENSG00000207087 |
. |
130268 |
0.05 |
| chr9_131085826_131085977 | 0.03 |
COQ4 |
coenzyme Q4 |
770 |
0.33 |
| chr16_47813458_47814031 | 0.03 |
PHKB |
phosphorylase kinase, beta |
83361 |
0.1 |
| chr6_151646770_151647383 | 0.03 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
253 |
0.88 |
| chr17_60868949_60869438 | 0.03 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
14808 |
0.23 |
| chr4_19778605_19778779 | 0.03 |
SLIT2 |
slit homolog 2 (Drosophila) |
476191 |
0.01 |
| chr14_38091284_38091435 | 0.03 |
TTC6 |
tetratricopeptide repeat domain 6 |
89 |
0.98 |
| chr8_39939130_39939407 | 0.03 |
C8orf4 |
chromosome 8 open reading frame 4 |
71721 |
0.11 |
| chr10_92794688_92794839 | 0.03 |
ENSG00000201604 |
. |
112171 |
0.06 |
| chr7_116141197_116141631 | 0.03 |
CAV2 |
caveolin 2 |
1580 |
0.29 |
| chr2_238186522_238186673 | 0.03 |
AC112715.2 |
Uncharacterized protein |
20863 |
0.24 |
| chr4_41157110_41157449 | 0.03 |
ENSG00000207198 |
. |
41320 |
0.14 |
| chr12_65702776_65702927 | 0.03 |
MSRB3 |
methionine sulfoxide reductase B3 |
17804 |
0.22 |
| chr3_184406321_184406472 | 0.03 |
MAGEF1 |
melanoma antigen family F, 1 |
23440 |
0.22 |
| chr10_29980674_29981313 | 0.03 |
ENSG00000222092 |
. |
19851 |
0.22 |
| chr17_53347750_53347901 | 0.03 |
HLF |
hepatic leukemia factor |
2648 |
0.31 |
| chr5_38783776_38783927 | 0.03 |
RP11-122C5.3 |
|
169 |
0.97 |
| chrX_46772292_46772991 | 0.03 |
JADE3 |
jade family PHD finger 3 |
410 |
0.86 |
| chr17_79197162_79197682 | 0.03 |
AZI1 |
5-azacytidine induced 1 |
623 |
0.55 |
| chr20_9141897_9142059 | 0.03 |
PLCB4 |
phospholipase C, beta 4 |
5027 |
0.33 |
| chr1_204010490_204010911 | 0.03 |
SOX13 |
SRY (sex determining region Y)-box 13 |
32032 |
0.15 |
| chr8_97596791_97596942 | 0.03 |
SDC2 |
syndecan 2 |
307 |
0.94 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0052308 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |