Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for HSF4
Z-value: 1.01
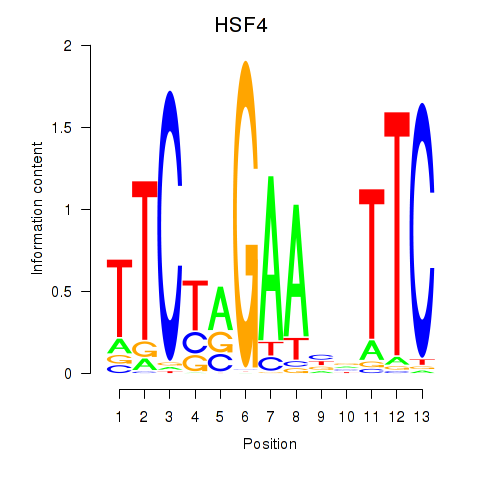
Transcription factors associated with HSF4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF4
|
ENSG00000102878.11 | heat shock transcription factor 4 |
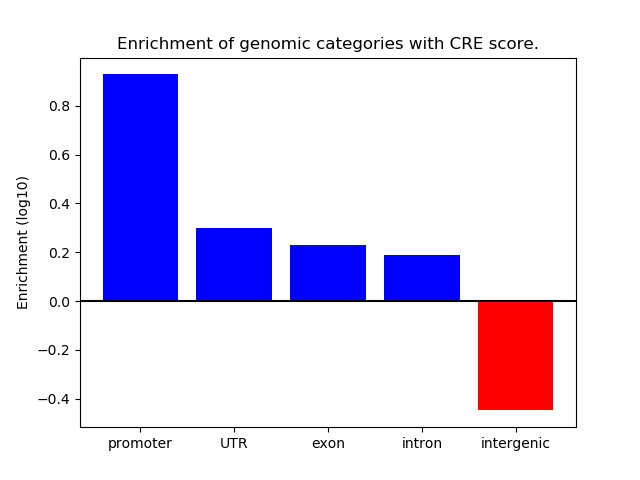
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_67196775_67197002 | HSF4 | 400 | 0.593118 | -0.69 | 4.1e-02 | Click! |
| chr16_67198733_67198897 | HSF4 | 100 | 0.901692 | -0.66 | 5.5e-02 | Click! |
| chr16_67199121_67199272 | HSF4 | 232 | 0.798377 | -0.58 | 1.0e-01 | Click! |
| chr16_67197260_67198336 | HSF4 | 510 | 0.518764 | -0.11 | 7.7e-01 | Click! |
Activity of the HSF4 motif across conditions
Conditions sorted by the z-value of the HSF4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
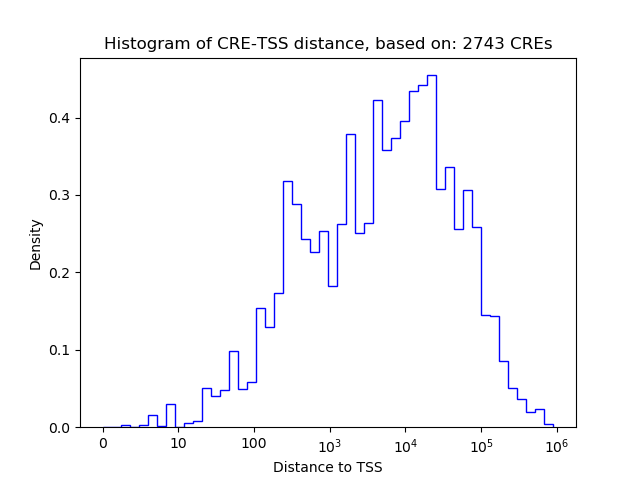
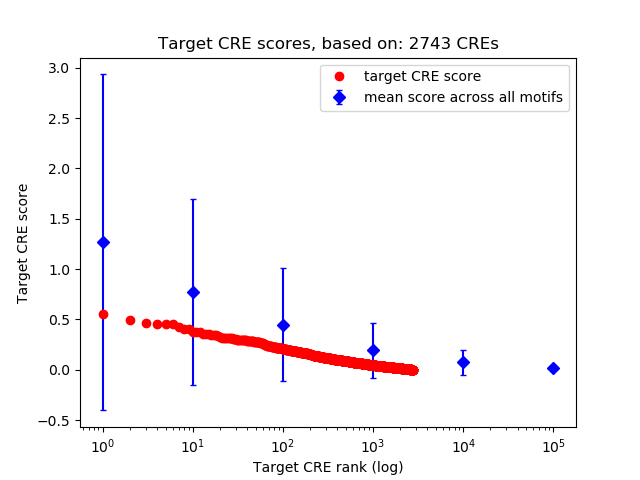
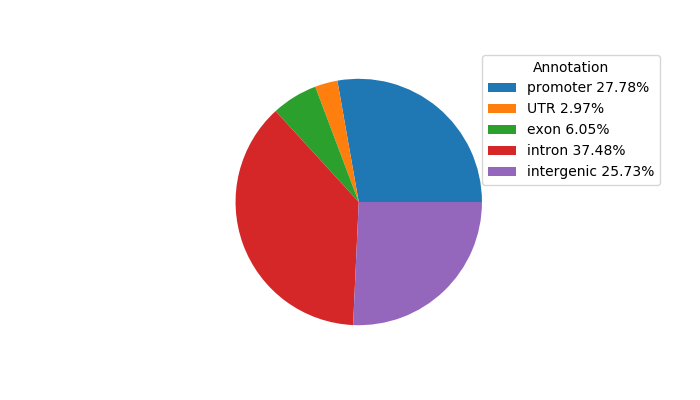
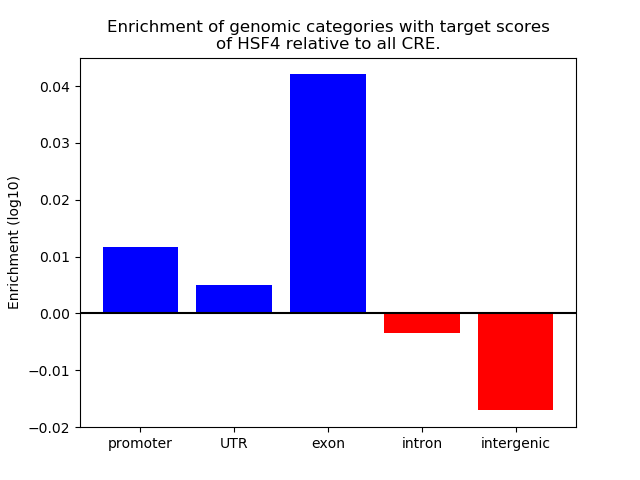
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_16493544_16493695 | 0.55 |
ENSG00000243745 |
. |
16151 |
0.14 |
| chrX_12968759_12969060 | 0.49 |
TMSB4X |
thymosin beta 4, X-linked |
24318 |
0.17 |
| chrY_7158530_7158855 | 0.47 |
PRKY |
protein kinase, Y-linked, pseudogene |
16338 |
0.2 |
| chr1_207997059_207997359 | 0.45 |
ENSG00000203709 |
. |
21341 |
0.22 |
| chrX_12925475_12925626 | 0.45 |
TLR8 |
toll-like receptor 8 |
792 |
0.45 |
| chr14_81425506_81425657 | 0.45 |
CEP128 |
centrosomal protein 128kDa |
247 |
0.94 |
| chrY_15815919_15816489 | 0.43 |
TMSB4Y |
thymosin beta 4, Y-linked |
757 |
0.72 |
| chr16_84628728_84628899 | 0.41 |
RP11-61F12.1 |
|
814 |
0.61 |
| chr1_145454279_145454446 | 0.40 |
TXNIP |
thioredoxin interacting protein |
15056 |
0.1 |
| chr16_79320926_79321088 | 0.38 |
ENSG00000222244 |
. |
22656 |
0.27 |
| chr20_48183167_48183318 | 0.38 |
PTGIS |
prostaglandin I2 (prostacyclin) synthase |
1441 |
0.45 |
| chr16_84582211_84582362 | 0.37 |
TLDC1 |
TBC/LysM-associated domain containing 1 |
5353 |
0.2 |
| chr17_45812399_45812645 | 0.35 |
TBX21 |
T-box 21 |
1912 |
0.27 |
| chr1_164695126_164695277 | 0.35 |
RP11-477H21.2 |
|
45879 |
0.13 |
| chr2_85936608_85936759 | 0.35 |
GNLY |
granulysin |
14192 |
0.13 |
| chr19_50003533_50003936 | 0.35 |
hsa-mir-150 |
hsa-mir-150 |
47 |
0.8 |
| chr2_176344322_176344473 | 0.34 |
ENSG00000221347 |
. |
149296 |
0.04 |
| chrX_123094531_123095374 | 0.34 |
STAG2 |
stromal antigen 2 |
212 |
0.95 |
| chr6_57043038_57043189 | 0.33 |
RP11-203B9.4 |
|
4079 |
0.22 |
| chr6_30459024_30459175 | 0.33 |
HLA-E |
major histocompatibility complex, class I, E |
1855 |
0.27 |
| chr3_95746418_95746569 | 0.32 |
ENSG00000221477 |
. |
139253 |
0.05 |
| chr19_16438433_16439148 | 0.32 |
KLF2 |
Kruppel-like factor 2 |
3139 |
0.19 |
| chr6_31323146_31323355 | 0.31 |
HLA-B |
major histocompatibility complex, class I, B |
1714 |
0.23 |
| chr5_72485454_72485605 | 0.31 |
TMEM174 |
transmembrane protein 174 |
16507 |
0.18 |
| chr11_122933023_122933174 | 0.31 |
HSPA8 |
heat shock 70kDa protein 8 |
38 |
0.96 |
| chrX_41777222_41777373 | 0.31 |
ENSG00000251807 |
. |
1305 |
0.5 |
| chr1_25246548_25246699 | 0.31 |
RUNX3 |
runt-related transcription factor 3 |
8989 |
0.23 |
| chr2_113545549_113545700 | 0.31 |
IL1A |
interleukin 1, alpha |
3457 |
0.21 |
| chr17_33863104_33863360 | 0.31 |
SLFN12L |
schlafen family member 12-like |
1648 |
0.24 |
| chrX_152989287_152989823 | 0.30 |
BCAP31 |
B-cell receptor-associated protein 31 |
6 |
0.92 |
| chr1_161493792_161494541 | 0.30 |
HSPA6 |
heat shock 70kDa protein 6 (HSP70B') |
130 |
0.94 |
| chr11_104788188_104788675 | 0.30 |
RP11-693N9.2 |
|
7789 |
0.2 |
| chr22_23528239_23528390 | 0.30 |
BCR |
breakpoint cluster region |
5762 |
0.16 |
| chr8_38323527_38324266 | 0.30 |
FGFR1 |
fibroblast growth factor receptor 1 |
285 |
0.89 |
| chr15_81593596_81593907 | 0.30 |
IL16 |
interleukin 16 |
1994 |
0.33 |
| chr9_21031432_21031950 | 0.30 |
PTPLAD2 |
protein tyrosine phosphatase-like A domain containing 2 |
56 |
0.98 |
| chr20_50148880_50149119 | 0.29 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
10259 |
0.28 |
| chr1_198629639_198630096 | 0.29 |
RP11-553K8.5 |
|
6323 |
0.26 |
| chr1_206718342_206718493 | 0.29 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
12076 |
0.16 |
| chr15_81576793_81576944 | 0.29 |
IL16 |
interleukin 16 |
12386 |
0.21 |
| chr4_111709350_111709501 | 0.29 |
ENSG00000251808 |
. |
65201 |
0.13 |
| chrX_55545866_55546017 | 0.29 |
USP51 |
ubiquitin specific peptidase 51 |
30306 |
0.17 |
| chr17_74137094_74137375 | 0.29 |
FOXJ1 |
forkhead box J1 |
146 |
0.74 |
| chr12_122228062_122228376 | 0.29 |
RHOF |
ras homolog family member F (in filopodia) |
3047 |
0.21 |
| chr22_37178306_37178457 | 0.29 |
IFT27 |
intraflagellar transport 27 homolog (Chlamydomonas) |
6184 |
0.17 |
| chr1_101187693_101187844 | 0.28 |
VCAM1 |
vascular cell adhesion molecule 1 |
2448 |
0.39 |
| chr19_17958522_17959083 | 0.28 |
JAK3 |
Janus kinase 3 |
24 |
0.96 |
| chr2_69003816_69004024 | 0.28 |
ARHGAP25 |
Rho GTPase activating protein 25 |
1848 |
0.42 |
| chr6_167566709_167567175 | 0.28 |
TCP10L2 |
t-complex 10-like 2 |
3974 |
0.21 |
| chr1_108484043_108484194 | 0.28 |
VAV3-AS1 |
VAV3 antisense RNA 1 |
22947 |
0.2 |
| chrX_15767915_15768266 | 0.27 |
CA5B |
carbonic anhydrase VB, mitochondrial |
3 |
0.98 |
| chr17_29877127_29877670 | 0.27 |
ENSG00000207614 |
. |
9617 |
0.11 |
| chr12_2903621_2904135 | 0.27 |
FKBP4 |
FK506 binding protein 4, 59kDa |
241 |
0.88 |
| chr9_110975706_110975857 | 0.27 |
ENSG00000222512 |
. |
145428 |
0.05 |
| chrX_70502857_70503125 | 0.27 |
NONO |
non-POU domain containing, octamer-binding |
51 |
0.97 |
| chr19_19728555_19728718 | 0.27 |
PBX4 |
pre-B-cell leukemia homeobox 4 |
1089 |
0.37 |
| chr7_50350295_50350528 | 0.27 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
2093 |
0.45 |
| chrX_133687659_133687810 | 0.27 |
ENSG00000223749 |
. |
6993 |
0.12 |
| chr2_69470356_69470507 | 0.27 |
ENSG00000199460 |
. |
60422 |
0.11 |
| chr4_7829564_7829715 | 0.27 |
AFAP1 |
actin filament associated protein 1 |
44170 |
0.15 |
| chr7_155283292_155284101 | 0.26 |
AC008060.5 |
|
19282 |
0.17 |
| chr14_22982783_22983297 | 0.25 |
TRAJ15 |
T cell receptor alpha joining 15 |
15540 |
0.1 |
| chr18_77165970_77166171 | 0.25 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
5678 |
0.26 |
| chr7_3083968_3084119 | 0.25 |
CARD11 |
caspase recruitment domain family, member 11 |
464 |
0.86 |
| chr9_78508950_78509101 | 0.25 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
3404 |
0.31 |
| chr8_23313563_23313898 | 0.24 |
ENTPD4 |
ectonucleoside triphosphate diphosphohydrolase 4 |
1430 |
0.42 |
| chr19_54875923_54876565 | 0.24 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
170 |
0.91 |
| chr1_145454503_145454819 | 0.24 |
TXNIP |
thioredoxin interacting protein |
15355 |
0.1 |
| chr1_54009379_54009530 | 0.23 |
DMRTB1 |
DMRT-like family B with proline-rich C-terminal, 1 |
84382 |
0.08 |
| chr10_95262845_95263083 | 0.23 |
CEP55 |
centrosomal protein 55kDa |
3803 |
0.2 |
| chr20_57722405_57722705 | 0.23 |
ZNF831 |
zinc finger protein 831 |
43520 |
0.15 |
| chr11_20384960_20385960 | 0.23 |
HTATIP2 |
HIV-1 Tat interactive protein 2, 30kDa |
60 |
0.98 |
| chr15_42837275_42837426 | 0.23 |
LRRC57 |
leucine rich repeat containing 57 |
3620 |
0.15 |
| chr17_8820194_8820345 | 0.23 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
4435 |
0.27 |
| chr19_55014108_55014308 | 0.23 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
85 |
0.94 |
| chr9_34988963_34989411 | 0.23 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
451 |
0.77 |
| chr1_119627265_119627416 | 0.23 |
WARS2-IT1 |
WARS2 intronic transcript 1 (non-protein coding) |
37312 |
0.16 |
| chr9_93914044_93914195 | 0.23 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
210058 |
0.03 |
| chr3_79625949_79626100 | 0.23 |
ENSG00000216116 |
. |
7810 |
0.3 |
| chr6_130992857_130993008 | 0.22 |
ENSG00000202438 |
. |
97675 |
0.08 |
| chr1_1711747_1711898 | 0.22 |
NADK |
NAD kinase |
314 |
0.86 |
| chr13_110951342_110951493 | 0.22 |
COL4A2 |
collagen, type IV, alpha 2 |
6742 |
0.23 |
| chr7_14019556_14019707 | 0.22 |
ETV1 |
ets variant 1 |
6435 |
0.28 |
| chrY_21906956_21907224 | 0.22 |
KDM5D |
lysine (K)-specific demethylase 5D |
265 |
0.96 |
| chr15_63789062_63789213 | 0.22 |
USP3 |
ubiquitin specific peptidase 3 |
7656 |
0.25 |
| chr9_132477050_132477230 | 0.22 |
RP11-483H20.6 |
|
3774 |
0.19 |
| chr4_26300916_26301067 | 0.22 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
20293 |
0.28 |
| chr8_56919486_56919637 | 0.22 |
ENSG00000241997 |
. |
15311 |
0.15 |
| chr15_101212076_101212240 | 0.22 |
ENSG00000212306 |
. |
33590 |
0.16 |
| chr3_71779214_71779365 | 0.22 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
941 |
0.61 |
| chr1_66797787_66798583 | 0.22 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
313 |
0.94 |
| chr6_14109075_14109337 | 0.21 |
CD83 |
CD83 molecule |
8666 |
0.27 |
| chr1_206753637_206754165 | 0.21 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
23408 |
0.13 |
| chr2_198651086_198651566 | 0.21 |
BOLL |
boule-like RNA-binding protein |
28 |
0.98 |
| chr8_123618804_123618955 | 0.21 |
ENSG00000238901 |
. |
64651 |
0.14 |
| chr3_119421432_119421650 | 0.21 |
MAATS1 |
MYCBP-associated, testis expressed 1 |
328 |
0.87 |
| chr9_139433085_139433378 | 0.21 |
RP11-413M3.4 |
|
4102 |
0.11 |
| chr14_72052659_72052844 | 0.21 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
247 |
0.96 |
| chr19_49164249_49164514 | 0.21 |
NTN5 |
netrin 5 |
11883 |
0.08 |
| chr15_63341000_63341752 | 0.21 |
RP11-244F12.3 |
|
71 |
0.83 |
| chr2_177361445_177361596 | 0.21 |
ENSG00000221304 |
. |
113216 |
0.06 |
| chr2_33391520_33391792 | 0.21 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
31932 |
0.22 |
| chr1_2163627_2164198 | 0.21 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
3778 |
0.16 |
| chr13_99966619_99966770 | 0.20 |
GPR183 |
G protein-coupled receptor 183 |
7035 |
0.21 |
| chr5_141737086_141737313 | 0.20 |
AC005592.2 |
|
4091 |
0.29 |
| chr19_18415340_18416257 | 0.20 |
LSM4 |
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
11271 |
0.09 |
| chr11_65837004_65837255 | 0.20 |
RP11-1167A19.2 |
Uncharacterized protein |
39 |
0.92 |
| chr20_30777439_30777911 | 0.20 |
PLAGL2 |
pleiomorphic adenoma gene-like 2 |
17919 |
0.14 |
| chr1_212731391_212732317 | 0.20 |
ATF3 |
activating transcription factor 3 |
6822 |
0.21 |
| chr19_18627573_18627724 | 0.20 |
ELL |
elongation factor RNA polymerase II |
5289 |
0.1 |
| chr22_24837615_24837766 | 0.20 |
ADORA2A |
adenosine A2a receptor |
8882 |
0.18 |
| chr4_1194452_1194845 | 0.20 |
SPON2 |
spondin 2, extracellular matrix protein |
4228 |
0.16 |
| chr11_48042023_48042428 | 0.20 |
AC103828.1 |
|
4818 |
0.24 |
| chr9_19441802_19441976 | 0.20 |
RP11-363E7.4 |
|
11318 |
0.18 |
| chr5_52021818_52021969 | 0.20 |
ITGA1 |
integrin, alpha 1 |
61837 |
0.12 |
| chr3_113252540_113252793 | 0.20 |
SIDT1 |
SID1 transmembrane family, member 1 |
1448 |
0.4 |
| chr19_50423027_50423178 | 0.20 |
NUP62 |
nucleoporin 62kDa |
2184 |
0.12 |
| chr19_4915707_4915936 | 0.20 |
ARRDC5 |
arrestin domain containing 5 |
12942 |
0.12 |
| chr3_72150163_72150348 | 0.20 |
ENSG00000212070 |
. |
161324 |
0.04 |
| chr6_45462799_45462992 | 0.19 |
RUNX2 |
runt-related transcription factor 2 |
72673 |
0.11 |
| chr6_29797302_29797453 | 0.19 |
HLA-G |
major histocompatibility complex, class I, G |
1755 |
0.34 |
| chr2_135052691_135052842 | 0.19 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
40936 |
0.19 |
| chr14_22694848_22695133 | 0.19 |
ENSG00000238634 |
. |
84103 |
0.09 |
| chr11_117821485_117821636 | 0.19 |
TMPRSS13 |
transmembrane protease, serine 13 |
21386 |
0.15 |
| chr16_1019853_1020655 | 0.19 |
LMF1 |
lipase maturation factor 1 |
612 |
0.62 |
| chr12_124768084_124768235 | 0.19 |
FAM101A |
family with sequence similarity 101, member A |
5551 |
0.28 |
| chr19_10927021_10927186 | 0.19 |
ENSG00000207752 |
. |
1069 |
0.38 |
| chr19_47217120_47217717 | 0.19 |
PRKD2 |
protein kinase D2 |
138 |
0.91 |
| chrX_9622413_9622564 | 0.19 |
GPR143 |
G protein-coupled receptor 143 |
111516 |
0.06 |
| chr7_138791837_138792259 | 0.19 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
2052 |
0.35 |
| chr10_111838058_111838209 | 0.19 |
ADD3 |
adducin 3 (gamma) |
70411 |
0.1 |
| chr16_47813458_47814031 | 0.19 |
PHKB |
phosphorylase kinase, beta |
83361 |
0.1 |
| chr6_41906196_41906466 | 0.19 |
CCND3 |
cyclin D3 |
2133 |
0.22 |
| chr8_142237735_142237886 | 0.19 |
SLC45A4 |
solute carrier family 45, member 4 |
859 |
0.55 |
| chr19_859742_860228 | 0.18 |
CFD |
complement factor D (adipsin) |
295 |
0.77 |
| chr19_36389493_36389866 | 0.18 |
NFKBID |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta |
1873 |
0.17 |
| chr1_9714142_9714379 | 0.18 |
C1orf200 |
chromosome 1 open reading frame 200 |
384 |
0.82 |
| chr10_104913694_104913845 | 0.18 |
NT5C2 |
5'-nucleotidase, cytosolic II |
385 |
0.89 |
| chr18_66469346_66469497 | 0.18 |
CCDC102B |
coiled-coil domain containing 102B |
3769 |
0.28 |
| chr18_6627846_6627997 | 0.18 |
ENSG00000243591 |
. |
26382 |
0.19 |
| chr12_58235429_58235580 | 0.18 |
CTDSP2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
3813 |
0.11 |
| chr19_2049519_2050886 | 0.18 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
650 |
0.55 |
| chr1_203279239_203279390 | 0.18 |
BTG2 |
BTG family, member 2 |
4650 |
0.19 |
| chr2_182245315_182245466 | 0.18 |
ENSG00000266705 |
. |
75011 |
0.11 |
| chr10_11212951_11213685 | 0.18 |
RP3-323N1.2 |
|
21 |
0.98 |
| chr6_112082790_112082941 | 0.18 |
FYN |
FYN oncogene related to SRC, FGR, YES |
1745 |
0.48 |
| chr3_130648074_130648225 | 0.18 |
RP11-39E3.3 |
|
671 |
0.43 |
| chrX_54071307_54071816 | 0.18 |
PHF8 |
PHD finger protein 8 |
121 |
0.97 |
| chrX_12995326_12995601 | 0.18 |
TMSB4X |
thymosin beta 4, X-linked |
1686 |
0.43 |
| chr7_46644240_46644391 | 0.18 |
AC011294.3 |
Uncharacterized protein |
92405 |
0.1 |
| chr15_28412952_28413117 | 0.18 |
HERC2 |
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
6535 |
0.28 |
| chr17_80398389_80398540 | 0.18 |
HEXDC |
hexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing |
3859 |
0.1 |
| chr19_17515989_17516450 | 0.18 |
BST2 |
bone marrow stromal cell antigen 2 |
238 |
0.54 |
| chr12_117298021_117298350 | 0.17 |
HRK |
harakiri, BCL2 interacting protein (contains only BH3 domain) |
20708 |
0.2 |
| chr2_100881771_100881922 | 0.17 |
LONRF2 |
LON peptidase N-terminal domain and ring finger 2 |
44121 |
0.17 |
| chrX_15768764_15768915 | 0.17 |
CA5B |
carbonic anhydrase VB, mitochondrial |
746 |
0.66 |
| chr1_202895737_202896386 | 0.17 |
KLHL12 |
kelch-like family member 12 |
329 |
0.82 |
| chr3_138893153_138893304 | 0.17 |
MRPS22 |
mitochondrial ribosomal protein S22 |
63982 |
0.12 |
| chr14_99699360_99699511 | 0.17 |
AL109767.1 |
|
29850 |
0.18 |
| chr2_101541390_101541618 | 0.17 |
NPAS2 |
neuronal PAS domain protein 2 |
104 |
0.97 |
| chr15_38855839_38856970 | 0.17 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
432 |
0.84 |
| chr5_100182433_100182584 | 0.17 |
ENSG00000221263 |
. |
30239 |
0.22 |
| chr2_65284469_65284714 | 0.17 |
CEP68 |
centrosomal protein 68kDa |
992 |
0.53 |
| chr1_158800341_158800492 | 0.17 |
MNDA |
myeloid cell nuclear differentiation antigen |
691 |
0.7 |
| chr8_28269075_28269620 | 0.17 |
ENSG00000207361 |
. |
4618 |
0.15 |
| chrX_153360358_153360509 | 0.17 |
MECP2 |
methyl CpG binding protein 2 (Rett syndrome) |
2003 |
0.26 |
| chr5_79331612_79331763 | 0.17 |
THBS4 |
thrombospondin 4 |
159 |
0.96 |
| chr19_18414393_18414628 | 0.17 |
LSM4 |
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
12559 |
0.09 |
| chr8_41593946_41594097 | 0.17 |
ANK1 |
ankyrin 1, erythrocytic |
30324 |
0.15 |
| chr10_117577321_117577472 | 0.17 |
GFRA1 |
GDNF family receptor alpha 1 |
437911 |
0.01 |
| chr9_132220609_132220886 | 0.17 |
ENSG00000264298 |
. |
20088 |
0.2 |
| chr2_144008418_144008569 | 0.17 |
RP11-190J23.1 |
|
78752 |
0.11 |
| chr12_125399791_125400355 | 0.17 |
ENSG00000265345 |
. |
20 |
0.69 |
| chr21_34431477_34431628 | 0.17 |
OLIG1 |
oligodendrocyte transcription factor 1 |
10898 |
0.17 |
| chr1_61637615_61637766 | 0.17 |
RP4-802A10.1 |
|
47285 |
0.17 |
| chr12_54688354_54688505 | 0.17 |
NFE2 |
nuclear factor, erythroid 2 |
1113 |
0.27 |
| chr17_63025367_63025525 | 0.17 |
RP11-583F2.5 |
|
22581 |
0.15 |
| chr2_113599646_113599797 | 0.17 |
ENSG00000221541 |
. |
2167 |
0.27 |
| chr1_93508255_93508475 | 0.17 |
ENSG00000251837 |
. |
15302 |
0.13 |
| chr14_99719284_99719435 | 0.17 |
AL109767.1 |
|
9926 |
0.22 |
| chrX_12990488_12990859 | 0.17 |
TMSB4X |
thymosin beta 4, X-linked |
2554 |
0.33 |
| chr8_19371412_19371563 | 0.16 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
87889 |
0.1 |
| chr1_27153904_27154578 | 0.16 |
ZDHHC18 |
zinc finger, DHHC-type containing 18 |
1040 |
0.39 |
| chr3_30852560_30852711 | 0.16 |
GADL1 |
glutamate decarboxylase-like 1 |
83518 |
0.1 |
| chr11_59537081_59537232 | 0.16 |
STX3 |
syntaxin 3 |
14231 |
0.13 |
| chr9_129183867_129184018 | 0.16 |
ENSG00000252985 |
. |
5230 |
0.22 |
| chr9_470290_470809 | 0.16 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
258 |
0.49 |
| chr21_44394961_44395127 | 0.16 |
PKNOX1 |
PBX/knotted 1 homeobox 1 |
293 |
0.89 |
| chr5_151098480_151098631 | 0.16 |
ENSG00000222102 |
. |
14416 |
0.14 |
| chr2_175458554_175459187 | 0.16 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
3623 |
0.22 |
| chr5_127577032_127577183 | 0.16 |
FBN2 |
fibrillin 2 |
97776 |
0.08 |
| chr3_108542080_108542294 | 0.16 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
568 |
0.83 |
| chr19_11159843_11160026 | 0.16 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
9577 |
0.14 |
| chr17_34610642_34610793 | 0.16 |
CCL3L1 |
chemokine (C-C motif) ligand 3-like 1 |
15002 |
0.13 |
| chr1_226001552_226001703 | 0.16 |
EPHX1 |
epoxide hydrolase 1, microsomal (xenobiotic) |
3791 |
0.18 |
| chr4_84030444_84030764 | 0.16 |
PLAC8 |
placenta-specific 8 |
392 |
0.89 |
| chr1_57038997_57039148 | 0.15 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
6169 |
0.3 |
| chr4_41574676_41574848 | 0.15 |
LIMCH1 |
LIM and calponin homology domains 1 |
34571 |
0.21 |
| chr1_204332360_204332939 | 0.15 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
3605 |
0.22 |
| chr11_75272983_75274329 | 0.15 |
SERPINH1 |
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
216 |
0.91 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.3 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.3 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.1 | 0.2 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.1 | 0.4 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.1 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.2 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.0 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.0 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.3 | GO:0019674 | NAD metabolic process(GO:0019674) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.2 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.0 | GO:2000757 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.0 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) |
| 0.0 | 0.0 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |