Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for IKZF2
Z-value: 1.11
Transcription factors associated with IKZF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IKZF2
|
ENSG00000030419.12 | IKAROS family zinc finger 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_213969171_213969322 | IKZF2 | 44107 | 0.187180 | 0.63 | 6.8e-02 | Click! |
| chr2_213969618_213969769 | IKZF2 | 43660 | 0.188675 | 0.63 | 7.1e-02 | Click! |
| chr2_213983567_213983718 | IKZF2 | 29711 | 0.237098 | 0.57 | 1.1e-01 | Click! |
| chr2_213983796_213984145 | IKZF2 | 29383 | 0.238239 | 0.51 | 1.6e-01 | Click! |
| chr2_214016688_214017732 | IKZF2 | 59 | 0.986711 | 0.49 | 1.8e-01 | Click! |
Activity of the IKZF2 motif across conditions
Conditions sorted by the z-value of the IKZF2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
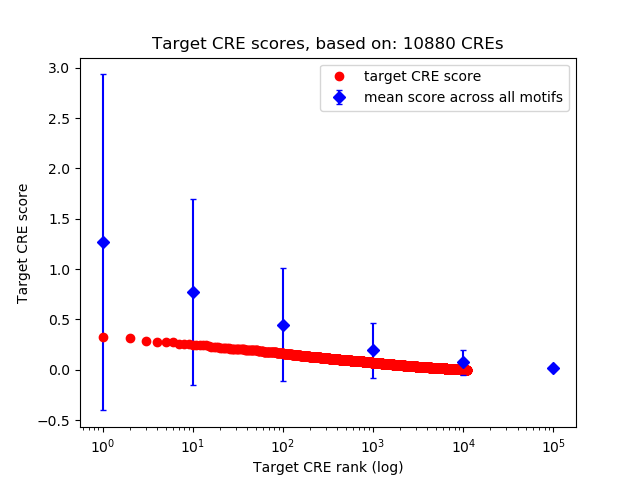
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_110720958_110721647 | 0.33 |
ATP2A2 |
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
1816 |
0.41 |
| chr12_116872831_116873001 | 0.31 |
ENSG00000264037 |
. |
6793 |
0.3 |
| chr3_29327656_29328192 | 0.28 |
RBMS3-AS3 |
RBMS3 antisense RNA 3 |
4293 |
0.26 |
| chr18_74277400_74278128 | 0.28 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
36890 |
0.17 |
| chr18_20558047_20558453 | 0.28 |
RBBP8 |
retinoblastoma binding protein 8 |
9445 |
0.15 |
| chr11_79861921_79862072 | 0.28 |
ENSG00000200146 |
. |
377008 |
0.01 |
| chr3_136649286_136649636 | 0.25 |
NCK1 |
NCK adaptor protein 1 |
117 |
0.97 |
| chr3_172240144_172241280 | 0.25 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
553 |
0.83 |
| chr3_155108862_155109013 | 0.25 |
ENSG00000272096 |
. |
46675 |
0.16 |
| chr9_34745557_34745708 | 0.25 |
FAM205A |
family with sequence similarity 205, member A |
16168 |
0.13 |
| chr3_5055440_5055591 | 0.25 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
33869 |
0.15 |
| chr12_32635081_32635232 | 0.25 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
3750 |
0.26 |
| chr20_11898508_11899014 | 0.24 |
BTBD3 |
BTB (POZ) domain containing 3 |
196 |
0.82 |
| chr14_103887907_103888058 | 0.24 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
35406 |
0.12 |
| chr7_93694115_93694266 | 0.24 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
60496 |
0.13 |
| chr21_39756340_39756646 | 0.23 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
87645 |
0.09 |
| chrX_73071645_73072457 | 0.23 |
RP13-216E22.5 |
|
117269 |
0.06 |
| chr10_21807539_21808360 | 0.23 |
SKIDA1 |
SKI/DACH domain containing 1 |
1101 |
0.4 |
| chr1_47489778_47489953 | 0.22 |
CYP4X1 |
cytochrome P450, family 4, subfamily X, polypeptide 1 |
625 |
0.75 |
| chr8_51783438_51783589 | 0.22 |
ENSG00000201316 |
. |
144068 |
0.05 |
| chr1_77751257_77751408 | 0.22 |
AK5 |
adenylate kinase 5 |
2568 |
0.34 |
| chr7_129255259_129255410 | 0.22 |
NRF1 |
nuclear respiratory factor 1 |
3737 |
0.29 |
| chr5_71806938_71807120 | 0.21 |
ZNF366 |
zinc finger protein 366 |
3475 |
0.33 |
| chr21_34415086_34415237 | 0.21 |
OLIG2 |
oligodendrocyte lineage transcription factor 2 |
16918 |
0.17 |
| chr18_19192929_19193396 | 0.21 |
SNRPD1 |
small nuclear ribonucleoprotein D1 polypeptide 16kDa |
870 |
0.52 |
| chr21_27944686_27945456 | 0.21 |
CYYR1 |
cysteine/tyrosine-rich 1 |
424 |
0.91 |
| chr21_16388120_16388271 | 0.21 |
NRIP1 |
nuclear receptor interacting protein 1 |
13506 |
0.22 |
| chr16_85160610_85160761 | 0.21 |
FAM92B |
family with sequence similarity 92, member B |
14571 |
0.21 |
| chr6_150083012_150083330 | 0.21 |
PCMT1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
11991 |
0.14 |
| chr1_9600473_9600668 | 0.21 |
SLC25A33 |
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
1029 |
0.58 |
| chr12_96839645_96840068 | 0.21 |
ENSG00000263364 |
. |
2598 |
0.28 |
| chr2_158694717_158695163 | 0.21 |
ACVR1 |
activin A receptor, type I |
101 |
0.97 |
| chr6_157104323_157104474 | 0.21 |
RP11-230C9.3 |
|
2809 |
0.25 |
| chr2_178032482_178032633 | 0.21 |
ENSG00000201102 |
. |
27154 |
0.12 |
| chr7_27957803_27957954 | 0.21 |
ENSG00000265382 |
. |
37772 |
0.18 |
| chr20_47448799_47448989 | 0.21 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
4474 |
0.31 |
| chr6_1314742_1314893 | 0.20 |
FOXQ1 |
forkhead box Q1 |
2142 |
0.4 |
| chr5_20109661_20109812 | 0.20 |
CDH18 |
cadherin 18, type 2 |
121397 |
0.07 |
| chr14_89673001_89673195 | 0.20 |
FOXN3 |
forkhead box N3 |
26010 |
0.24 |
| chr7_48675933_48676084 | 0.20 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
128540 |
0.06 |
| chr6_109414649_109415318 | 0.20 |
SESN1 |
sestrin 1 |
1039 |
0.42 |
| chr5_64588165_64588316 | 0.20 |
ENSG00000207439 |
. |
169044 |
0.03 |
| chr2_102466743_102466894 | 0.20 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
5621 |
0.28 |
| chr3_196697012_196697163 | 0.20 |
PIGZ |
phosphatidylinositol glycan anchor biosynthesis, class Z |
1345 |
0.35 |
| chr6_27247805_27248026 | 0.20 |
POM121L2 |
POM121 transmembrane nucleoporin-like 2 |
31176 |
0.15 |
| chr12_90287998_90288165 | 0.20 |
ENSG00000252823 |
. |
140245 |
0.05 |
| chr20_10399686_10399837 | 0.20 |
MKKS |
McKusick-Kaufman syndrome |
12809 |
0.19 |
| chr2_113591900_113592051 | 0.19 |
IL1B |
interleukin 1, beta |
2036 |
0.29 |
| chr15_43885389_43885540 | 0.19 |
CKMT1B |
creatine kinase, mitochondrial 1B |
170 |
0.9 |
| chr3_129383548_129383699 | 0.19 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
8054 |
0.22 |
| chr3_189515104_189515255 | 0.19 |
TP63 |
tumor protein p63 |
7589 |
0.27 |
| chr1_150481112_150481644 | 0.19 |
ECM1 |
extracellular matrix protein 1 |
795 |
0.48 |
| chr5_171389606_171389757 | 0.19 |
FBXW11 |
F-box and WD repeat domain containing 11 |
15079 |
0.25 |
| chr3_87000039_87000329 | 0.19 |
VGLL3 |
vestigial like 3 (Drosophila) |
39668 |
0.23 |
| chr21_17959283_17959568 | 0.19 |
ENSG00000207863 |
. |
3132 |
0.31 |
| chr2_64878829_64879050 | 0.19 |
SERTAD2 |
SERTA domain containing 2 |
2108 |
0.38 |
| chr12_50410240_50410391 | 0.18 |
RACGAP1 |
Rac GTPase activating protein 1 |
188 |
0.92 |
| chr3_42864733_42864884 | 0.18 |
ACKR2 |
atypical chemokine receptor 2 |
3697 |
0.14 |
| chr7_27211508_27211659 | 0.18 |
HOXA10 |
homeobox A10 |
2342 |
0.09 |
| chr7_30326052_30326514 | 0.18 |
ZNRF2 |
zinc and ring finger 2 |
2360 |
0.29 |
| chr2_192842795_192842946 | 0.18 |
AC098617.1 |
|
43768 |
0.2 |
| chr3_132036252_132036528 | 0.18 |
ACPP |
acid phosphatase, prostate |
139 |
0.97 |
| chr12_66198631_66198915 | 0.18 |
HMGA2 |
high mobility group AT-hook 2 |
19138 |
0.21 |
| chr9_90133589_90133740 | 0.18 |
DAPK1 |
death-associated protein kinase 1 |
19744 |
0.23 |
| chr2_172964285_172964474 | 0.18 |
DLX2 |
distal-less homeobox 2 |
3249 |
0.26 |
| chr2_9348603_9349207 | 0.18 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
2011 |
0.46 |
| chr9_90955493_90955644 | 0.18 |
ENSG00000252299 |
. |
33616 |
0.2 |
| chr17_57289227_57289378 | 0.18 |
SMG8 |
SMG8 nonsense mediated mRNA decay factor |
518 |
0.64 |
| chr17_56490979_56491130 | 0.18 |
RNF43 |
ring finger protein 43 |
1948 |
0.27 |
| chr12_13332071_13332496 | 0.18 |
EMP1 |
epithelial membrane protein 1 |
17367 |
0.23 |
| chr2_66803096_66803300 | 0.18 |
MEIS1 |
Meis homeobox 1 |
67139 |
0.13 |
| chr14_94255244_94255403 | 0.18 |
PRIMA1 |
proline rich membrane anchor 1 |
496 |
0.86 |
| chr6_86349087_86349642 | 0.18 |
SYNCRIP |
synaptotagmin binding, cytoplasmic RNA interacting protein |
3345 |
0.23 |
| chr12_104360447_104360701 | 0.18 |
TDG |
thymine-DNA glycosylase |
910 |
0.37 |
| chr3_71478030_71478538 | 0.18 |
ENSG00000221264 |
. |
112956 |
0.06 |
| chr19_31830586_31830803 | 0.18 |
AC007796.1 |
|
9093 |
0.25 |
| chr3_99619729_99619880 | 0.17 |
FILIP1L |
filamin A interacting protein 1-like |
24758 |
0.22 |
| chr20_61555717_61555868 | 0.17 |
DIDO1 |
death inducer-obliterator 1 |
2029 |
0.26 |
| chr11_67979557_67979856 | 0.17 |
SUV420H1 |
suppressor of variegation 4-20 homolog 1 (Drosophila) |
515 |
0.8 |
| chr10_108792061_108792212 | 0.17 |
ENSG00000200626 |
. |
62357 |
0.16 |
| chr15_90364678_90364977 | 0.17 |
ANPEP |
alanyl (membrane) aminopeptidase |
6194 |
0.14 |
| chr8_70745749_70746362 | 0.17 |
RP11-159H10.3 |
|
291 |
0.62 |
| chr6_99798162_99798313 | 0.17 |
FAXC |
failed axon connections homolog (Drosophila) |
706 |
0.71 |
| chr2_145438866_145439017 | 0.17 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
160320 |
0.04 |
| chr4_71388674_71388825 | 0.17 |
AMTN |
amelotin |
4447 |
0.22 |
| chr6_108886554_108887901 | 0.17 |
FOXO3 |
forkhead box O3 |
5158 |
0.32 |
| chr16_87469465_87469755 | 0.17 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
2870 |
0.22 |
| chr11_47615339_47615925 | 0.17 |
C1QTNF4 |
C1q and tumor necrosis factor related protein 4 |
579 |
0.54 |
| chr3_42135461_42135612 | 0.17 |
TRAK1 |
trafficking protein, kinesin binding 1 |
2974 |
0.36 |
| chr12_51422377_51422723 | 0.17 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
201 |
0.91 |
| chr1_28385256_28385469 | 0.17 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
764 |
0.61 |
| chr7_22861355_22861506 | 0.17 |
TOMM7 |
translocase of outer mitochondrial membrane 7 homolog (yeast) |
533 |
0.81 |
| chr5_51742403_51742554 | 0.17 |
ITGA1 |
integrin, alpha 1 |
341252 |
0.01 |
| chr18_24083261_24083412 | 0.17 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
43627 |
0.17 |
| chr10_118897625_118897901 | 0.17 |
VAX1 |
ventral anterior homeobox 1 |
49 |
0.97 |
| chr9_13278507_13279169 | 0.17 |
RP11-272P10.2 |
|
348 |
0.67 |
| chr12_48176505_48176789 | 0.17 |
AC004466.1 |
Uncharacterized protein |
2059 |
0.24 |
| chr15_98538729_98539013 | 0.17 |
ENSG00000252477 |
. |
30400 |
0.18 |
| chr6_65214990_65215141 | 0.17 |
RP11-349P19.1 |
|
127377 |
0.06 |
| chr8_38857241_38857490 | 0.16 |
ADAM9 |
ADAM metallopeptidase domain 9 |
2860 |
0.19 |
| chr8_117449236_117449387 | 0.16 |
ENSG00000264815 |
. |
149723 |
0.04 |
| chr1_157014914_157015379 | 0.16 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
16 |
0.98 |
| chr6_37136440_37136591 | 0.16 |
PIM1 |
pim-1 oncogene |
1464 |
0.41 |
| chr3_112931196_112931871 | 0.16 |
BOC |
BOC cell adhesion associated, oncogene regulated |
124 |
0.97 |
| chrX_73471805_73471956 | 0.16 |
ENSG00000265727 |
. |
9144 |
0.12 |
| chr1_214619591_214619742 | 0.16 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
18480 |
0.26 |
| chr20_14317720_14318261 | 0.16 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
264 |
0.94 |
| chr15_57373111_57373262 | 0.16 |
ENSG00000222586 |
. |
114591 |
0.06 |
| chr19_56166514_56167932 | 0.16 |
U2AF2 |
U2 small nuclear RNA auxiliary factor 2 |
792 |
0.34 |
| chr3_119807952_119808228 | 0.16 |
GSK3B |
glycogen synthase kinase 3 beta |
4423 |
0.23 |
| chr4_42153064_42153252 | 0.16 |
BEND4 |
BEN domain containing 4 |
1737 |
0.5 |
| chr9_2160189_2160438 | 0.16 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
368 |
0.87 |
| chr8_48572289_48573010 | 0.16 |
SPIDR |
scaffolding protein involved in DNA repair |
423 |
0.86 |
| chr5_112762315_112762466 | 0.16 |
CTD-2201G3.1 |
|
6885 |
0.21 |
| chr2_17009553_17009704 | 0.16 |
FAM49A |
family with sequence similarity 49, member A |
162029 |
0.04 |
| chr2_198464213_198464591 | 0.15 |
RFTN2 |
raftlin family member 2 |
18217 |
0.19 |
| chr15_43985226_43985377 | 0.15 |
CKMT1A |
creatine kinase, mitochondrial 1A |
203 |
0.87 |
| chr13_78659821_78660303 | 0.15 |
RNF219-AS1 |
RNF219 antisense RNA 1 |
30772 |
0.24 |
| chr3_171065824_171066073 | 0.15 |
ENSG00000222506 |
. |
91367 |
0.09 |
| chr19_11449502_11449733 | 0.15 |
RAB3D |
RAB3D, member RAS oncogene family |
727 |
0.43 |
| chr17_26369297_26369946 | 0.15 |
NLK |
nemo-like kinase |
244 |
0.91 |
| chr6_26184757_26184908 | 0.15 |
HIST1H2BE |
histone cluster 1, H2be |
874 |
0.29 |
| chr4_164253130_164253399 | 0.15 |
NPY1R |
neuropeptide Y receptor Y1 |
484 |
0.84 |
| chr1_67398755_67399224 | 0.15 |
MIER1 |
mesoderm induction early response 1, transcriptional regulator |
3063 |
0.27 |
| chr8_48770878_48771029 | 0.15 |
ENSG00000211562 |
. |
31666 |
0.17 |
| chr20_4415632_4415783 | 0.15 |
ADRA1D |
adrenoceptor alpha 1D |
185986 |
0.03 |
| chr2_186509945_186510096 | 0.15 |
ENSG00000212581 |
. |
16481 |
0.26 |
| chr1_34368788_34368939 | 0.15 |
RP5-1007G16.1 |
|
27796 |
0.2 |
| chr5_130066537_130066688 | 0.15 |
ENSG00000252514 |
. |
344239 |
0.01 |
| chr14_65566001_65566152 | 0.15 |
MAX |
MYC associated factor X |
2981 |
0.21 |
| chr20_44632803_44632954 | 0.15 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
4669 |
0.14 |
| chr7_79767051_79767278 | 0.15 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
2093 |
0.45 |
| chr1_86284190_86284341 | 0.15 |
ENSG00000201620 |
. |
65216 |
0.13 |
| chr14_35871775_35873043 | 0.15 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
937 |
0.68 |
| chr2_27631132_27631707 | 0.15 |
PPM1G |
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
1030 |
0.33 |
| chrX_16825753_16826181 | 0.15 |
TXLNG |
taxilin gamma |
21412 |
0.16 |
| chr15_39886218_39886369 | 0.15 |
CTD-2033D15.1 |
|
139 |
0.96 |
| chr13_27975562_27975713 | 0.15 |
ENSG00000201242 |
. |
1087 |
0.48 |
| chr7_93999537_93999688 | 0.15 |
COL1A2 |
collagen, type I, alpha 2 |
24261 |
0.25 |
| chr2_140023256_140023407 | 0.15 |
ENSG00000206901 |
. |
245663 |
0.02 |
| chr18_12315741_12315987 | 0.15 |
TUBB6 |
tubulin, beta 6 class V |
4898 |
0.19 |
| chr11_75210272_75210450 | 0.15 |
GDPD5 |
glycerophosphodiester phosphodiesterase domain containing 5 |
8565 |
0.15 |
| chr3_168864469_168864620 | 0.15 |
MECOM |
MDS1 and EVI1 complex locus |
81 |
0.99 |
| chr7_28724839_28725039 | 0.15 |
CREB5 |
cAMP responsive element binding protein 5 |
659 |
0.83 |
| chr15_91204059_91204503 | 0.14 |
RP11-387D10.3 |
|
311 |
0.88 |
| chr4_31024444_31024595 | 0.14 |
RP11-619J20.1 |
|
228927 |
0.02 |
| chr6_145124672_145124823 | 0.14 |
UTRN |
utrophin |
4230 |
0.38 |
| chr7_155090835_155091229 | 0.14 |
INSIG1 |
insulin induced gene 1 |
761 |
0.7 |
| chr6_137532057_137532208 | 0.14 |
IFNGR1 |
interferon gamma receptor 1 |
7519 |
0.26 |
| chr8_25070650_25071114 | 0.14 |
DOCK5 |
dedicator of cytokinesis 5 |
28502 |
0.2 |
| chr1_183156151_183156302 | 0.14 |
LAMC2 |
laminin, gamma 2 |
803 |
0.7 |
| chr10_77158203_77158354 | 0.14 |
ENSG00000237149 |
. |
2999 |
0.24 |
| chr17_26220981_26221377 | 0.14 |
LYRM9 |
LYR motif containing 9 |
770 |
0.4 |
| chr10_97154342_97154493 | 0.14 |
SORBS1 |
sorbin and SH3 domain containing 1 |
21035 |
0.25 |
| chr5_54605223_54605374 | 0.14 |
SKIV2L2 |
superkiller viralicidic activity 2-like 2 (S. cerevisiae) |
1469 |
0.33 |
| chr15_45933283_45933523 | 0.14 |
SQRDL |
sulfide quinone reductase-like (yeast) |
4676 |
0.21 |
| chr1_159061696_159061989 | 0.14 |
AIM2 |
absent in melanoma 2 |
15151 |
0.17 |
| chr2_60764234_60764468 | 0.14 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
16189 |
0.22 |
| chr3_193852851_193853388 | 0.14 |
HES1 |
hes family bHLH transcription factor 1 |
815 |
0.62 |
| chr4_143322143_143322556 | 0.14 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
30063 |
0.27 |
| chr15_67334158_67334332 | 0.14 |
SMAD3 |
SMAD family member 3 |
21856 |
0.25 |
| chr20_57431129_57431280 | 0.14 |
GNAS |
GNAS complex locus |
971 |
0.45 |
| chr11_130818088_130818239 | 0.14 |
SNX19 |
sorting nexin 19 |
31759 |
0.21 |
| chr4_187491209_187491867 | 0.14 |
MTNR1A |
melatonin receptor 1A |
14817 |
0.17 |
| chr2_190647319_190647470 | 0.14 |
ORMDL1 |
ORM1-like 1 (S. cerevisiae) |
714 |
0.49 |
| chr10_27815365_27815516 | 0.14 |
RAB18 |
RAB18, member RAS oncogene family |
21892 |
0.24 |
| chr15_59062577_59062767 | 0.14 |
FAM63B |
family with sequence similarity 63, member B |
841 |
0.54 |
| chr4_124426780_124427488 | 0.14 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
106011 |
0.08 |
| chr13_48796681_48797014 | 0.14 |
ITM2B |
integral membrane protein 2B |
10447 |
0.26 |
| chr5_76527145_76527311 | 0.14 |
PDE8B |
phosphodiesterase 8B |
17459 |
0.23 |
| chr1_174160405_174160556 | 0.14 |
ENSG00000238872 |
. |
8888 |
0.23 |
| chr1_231469833_231470794 | 0.14 |
SPRTN |
SprT-like N-terminal domain |
2537 |
0.25 |
| chr13_21000022_21000173 | 0.14 |
ENSG00000263978 |
. |
7888 |
0.27 |
| chr2_238732566_238732717 | 0.14 |
RBM44 |
RNA binding motif protein 44 |
24820 |
0.17 |
| chr2_144382037_144382188 | 0.14 |
AC092652.1 |
|
24342 |
0.17 |
| chr6_21597329_21597632 | 0.14 |
SOX4 |
SRY (sex determining region Y)-box 4 |
3409 |
0.4 |
| chr1_215742028_215742345 | 0.14 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
1451 |
0.56 |
| chr11_737225_737376 | 0.14 |
TALDO1 |
transaldolase 1 |
10029 |
0.07 |
| chr2_217859430_217859581 | 0.14 |
AC007563.1 |
|
120850 |
0.05 |
| chr1_151963774_151963995 | 0.14 |
S100A10 |
S100 calcium binding protein A10 |
1170 |
0.39 |
| chr8_12989527_12990464 | 0.14 |
DLC1 |
deleted in liver cancer 1 |
988 |
0.62 |
| chr1_109583292_109583606 | 0.14 |
WDR47 |
WD repeat domain 47 |
720 |
0.6 |
| chr1_161703743_161703894 | 0.14 |
ENSG00000241347 |
. |
1780 |
0.22 |
| chr15_38558851_38559240 | 0.14 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
13663 |
0.3 |
| chr2_175202799_175202950 | 0.14 |
AC018470.1 |
Uncharacterized protein FLJ46347 |
723 |
0.62 |
| chr12_29434762_29434913 | 0.13 |
RP11-996F15.2 |
|
35944 |
0.16 |
| chr8_66752673_66753180 | 0.13 |
PDE7A |
phosphodiesterase 7A |
817 |
0.75 |
| chr9_140524792_140524954 | 0.13 |
EHMT1 |
euchromatic histone-lysine N-methyltransferase 1 |
11419 |
0.13 |
| chr5_14036124_14036305 | 0.13 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
91562 |
0.09 |
| chr8_98928953_98929409 | 0.13 |
MATN2 |
matrilin 2 |
14388 |
0.23 |
| chr9_17136040_17136191 | 0.13 |
CNTLN |
centlein, centrosomal protein |
1077 |
0.67 |
| chr8_71359268_71359419 | 0.13 |
ENSG00000253143 |
. |
33379 |
0.17 |
| chr16_75033273_75033750 | 0.13 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
268 |
0.92 |
| chr6_122719971_122720474 | 0.13 |
HSF2 |
heat shock transcription factor 2 |
469 |
0.87 |
| chr7_74074778_74074955 | 0.13 |
GTF2I |
general transcription factor IIi |
2509 |
0.33 |
| chr20_45982305_45982568 | 0.13 |
RP4-569M23.2 |
|
1131 |
0.31 |
| chr4_159417737_159417888 | 0.13 |
RP11-781M16.2 |
|
13248 |
0.22 |
| chr16_19769260_19769411 | 0.13 |
CTD-2380F24.1 |
|
8031 |
0.18 |
| chr1_43151006_43151188 | 0.13 |
YBX1 |
Y box binding protein 1 |
2439 |
0.25 |
| chr5_39412000_39412151 | 0.13 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
12895 |
0.26 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.2 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.1 | 0.2 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.2 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.2 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.2 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.2 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:0032823 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0031659 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0070391 | response to lipoteichoic acid(GO:0070391) cellular response to lipoteichoic acid(GO:0071223) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.0 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0043552 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.2 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0032730 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0060573 | ventral spinal cord interneuron differentiation(GO:0021514) ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.0 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.0 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.1 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0098930 | axonal transport(GO:0098930) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.1 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.0 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.0 | GO:2000757 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.0 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.3 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.3 | GO:0019079 | viral genome replication(GO:0019079) |
| 0.0 | 0.0 | GO:0090224 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 0.0 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.3 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.4 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.0 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |