Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
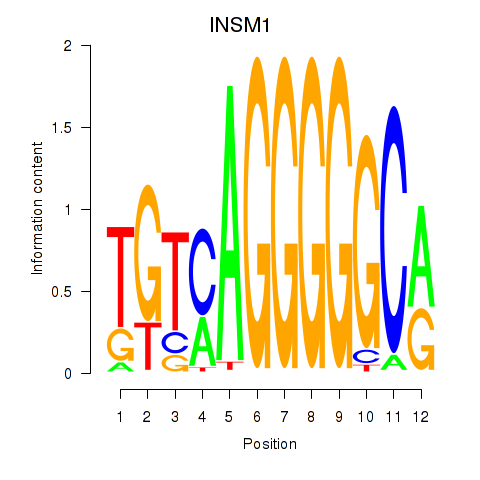
Results for INSM1
Z-value: 0.59
Transcription factors associated with INSM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
INSM1
|
ENSG00000173404.3 | INSM transcriptional repressor 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr20_20353422_20353573 | INSM1 | 4732 | 0.279002 | 0.49 | 1.8e-01 | Click! |
| chr20_20345257_20345408 | INSM1 | 3433 | 0.300789 | 0.33 | 3.9e-01 | Click! |
| chr20_20347903_20348069 | INSM1 | 779 | 0.720076 | -0.32 | 4.0e-01 | Click! |
| chr20_20348086_20348409 | INSM1 | 518 | 0.838818 | 0.31 | 4.2e-01 | Click! |
| chr20_20348603_20348876 | INSM1 | 26 | 0.984072 | 0.25 | 5.2e-01 | Click! |
Activity of the INSM1 motif across conditions
Conditions sorted by the z-value of the INSM1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
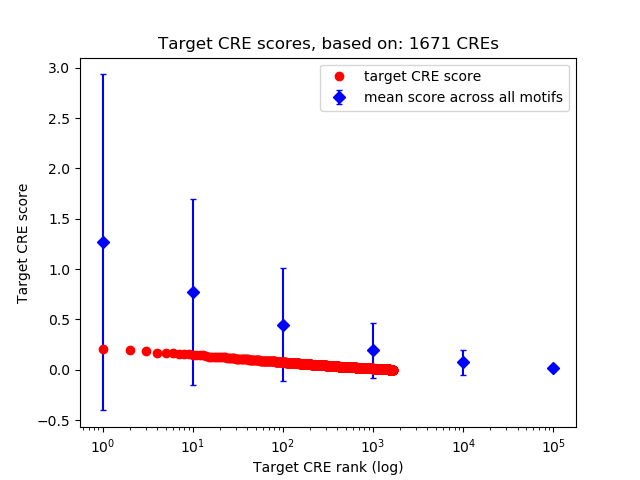
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_9545673_9545824 | 0.21 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
439 |
0.82 |
| chr19_3290313_3290464 | 0.19 |
AC010649.1 |
|
12531 |
0.17 |
| chr3_31238726_31238877 | 0.19 |
ENSG00000222983 |
. |
31406 |
0.2 |
| chr5_63933889_63934062 | 0.17 |
FAM159B |
family with sequence similarity 159, member B |
52160 |
0.14 |
| chr2_72374385_72374536 | 0.17 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
503 |
0.89 |
| chr8_99076485_99077142 | 0.17 |
C8orf47 |
chromosome 8 open reading frame 47 |
63 |
0.96 |
| chr5_139422603_139423172 | 0.16 |
NRG2 |
neuregulin 2 |
3 |
0.98 |
| chr1_177139870_177140730 | 0.16 |
BRINP2 |
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
333 |
0.89 |
| chr10_133775388_133775627 | 0.16 |
BNIP3 |
BCL2/adenovirus E1B 19kDa interacting protein 3 |
19920 |
0.21 |
| chr1_94701176_94701460 | 0.15 |
ARHGAP29 |
Rho GTPase activating protein 29 |
1803 |
0.43 |
| chr17_15164779_15165041 | 0.15 |
RP11-849N15.1 |
|
578 |
0.55 |
| chr12_67807670_67807821 | 0.15 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
114993 |
0.07 |
| chr1_19991906_19992120 | 0.15 |
HTR6 |
5-hydroxytryptamine (serotonin) receptor 6, G protein-coupled |
233 |
0.92 |
| chr4_148402008_148402467 | 0.13 |
EDNRA |
endothelin receptor type A |
153 |
0.97 |
| chr22_47075931_47076082 | 0.13 |
GRAMD4 |
GRAM domain containing 4 |
5501 |
0.25 |
| chr7_2528109_2528260 | 0.13 |
LFNG |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
23979 |
0.15 |
| chr11_70244216_70245100 | 0.13 |
CTTN |
cortactin |
11 |
0.53 |
| chr1_201436884_201437966 | 0.13 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
887 |
0.55 |
| chr2_177022446_177022597 | 0.13 |
HOXD3 |
homeobox D3 |
3098 |
0.11 |
| chr10_89353641_89353792 | 0.13 |
ENSG00000222192 |
. |
30773 |
0.17 |
| chr1_1280492_1280843 | 0.13 |
DVL1 |
dishevelled segment polarity protein 1 |
3825 |
0.08 |
| chr1_164531809_164532055 | 0.13 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
110 |
0.98 |
| chr19_13113568_13113719 | 0.13 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
6991 |
0.09 |
| chr16_21274943_21275094 | 0.12 |
CRYM |
crystallin, mu |
1539 |
0.38 |
| chr7_116165094_116165543 | 0.12 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
263 |
0.9 |
| chrX_153770529_153770697 | 0.12 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
135 |
0.91 |
| chr1_27830414_27830820 | 0.12 |
WASF2 |
WAS protein family, member 2 |
13948 |
0.16 |
| chr7_100464814_100466140 | 0.11 |
TRIP6 |
thyroid hormone receptor interactor 6 |
717 |
0.47 |
| chr19_13136463_13136690 | 0.11 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
765 |
0.47 |
| chr17_40263956_40264570 | 0.11 |
DHX58 |
DEXH (Asp-Glu-X-His) box polypeptide 58 |
218 |
0.87 |
| chr7_36343576_36343802 | 0.11 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
6936 |
0.18 |
| chr2_236403404_236403896 | 0.11 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
899 |
0.65 |
| chr8_145045264_145045480 | 0.11 |
PLEC |
plectin |
2325 |
0.16 |
| chr15_74044526_74044677 | 0.11 |
C15orf59 |
chromosome 15 open reading frame 59 |
118 |
0.97 |
| chr17_79438988_79439155 | 0.11 |
RP11-1055B8.8 |
|
10108 |
0.1 |
| chr11_12309017_12309498 | 0.11 |
MICALCL |
MICAL C-terminal like |
810 |
0.62 |
| chr8_74332619_74332912 | 0.11 |
RP11-434I12.2 |
|
64069 |
0.13 |
| chr19_13110560_13110829 | 0.10 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
4042 |
0.11 |
| chr15_96888963_96889465 | 0.10 |
ENSG00000222651 |
. |
12724 |
0.15 |
| chr5_148788814_148788965 | 0.10 |
ENSG00000208035 |
. |
19592 |
0.11 |
| chr19_34973190_34973467 | 0.10 |
WTIP |
Wilms tumor 1 interacting protein |
105 |
0.96 |
| chr2_66808823_66809387 | 0.10 |
MEIS1 |
Meis homeobox 1 |
73046 |
0.12 |
| chr15_96903910_96904291 | 0.10 |
AC087477.1 |
Uncharacterized protein |
387 |
0.85 |
| chr1_202163240_202163463 | 0.10 |
LGR6 |
leucine-rich repeat containing G protein-coupled receptor 6 |
322 |
0.85 |
| chr17_37755031_37755259 | 0.10 |
NEUROD2 |
neuronal differentiation 2 |
9051 |
0.14 |
| chr9_130659943_130660240 | 0.10 |
ST6GALNAC6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
200 |
0.87 |
| chr4_1165420_1165733 | 0.10 |
SPON2 |
spondin 2, extracellular matrix protein |
1085 |
0.38 |
| chr19_8431934_8432257 | 0.10 |
ANGPTL4 |
angiopoietin-like 4 |
2813 |
0.13 |
| chr17_39472322_39472473 | 0.10 |
KRTAP17-1 |
keratin associated protein 17-1 |
450 |
0.64 |
| chr6_72892413_72892934 | 0.10 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
218 |
0.96 |
| chr15_75943747_75944181 | 0.10 |
SNX33 |
sorting nexin 33 |
1867 |
0.18 |
| chr9_107622486_107622637 | 0.10 |
RP11-217B7.2 |
|
67273 |
0.1 |
| chr2_74666737_74667617 | 0.09 |
RTKN |
rhotekin |
533 |
0.54 |
| chr11_69453209_69453537 | 0.09 |
CCND1 |
cyclin D1 |
2482 |
0.32 |
| chr10_12631674_12631825 | 0.09 |
ENSG00000263584 |
. |
10997 |
0.27 |
| chr12_57522507_57523183 | 0.09 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
38 |
0.64 |
| chr22_43547615_43548743 | 0.09 |
TSPO |
translocator protein (18kDa) |
242 |
0.9 |
| chr11_64001151_64001876 | 0.09 |
RP11-783K16.14 |
|
251 |
0.61 |
| chr8_142010910_142011208 | 0.09 |
PTK2 |
protein tyrosine kinase 2 |
23 |
0.98 |
| chr17_13505362_13505866 | 0.09 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
370 |
0.91 |
| chr7_100765722_100766153 | 0.09 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
4433 |
0.12 |
| chr6_168435834_168435985 | 0.09 |
KIF25 |
kinesin family member 25 |
1231 |
0.55 |
| chr17_47574421_47575220 | 0.09 |
NGFR |
nerve growth factor receptor |
67 |
0.97 |
| chr8_22241251_22241835 | 0.09 |
SLC39A14 |
solute carrier family 39 (zinc transporter), member 14 |
8800 |
0.18 |
| chr19_6740138_6741070 | 0.09 |
TRIP10 |
thyroid hormone receptor interactor 10 |
325 |
0.81 |
| chr13_102105916_102106117 | 0.09 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
998 |
0.66 |
| chr7_75896724_75896875 | 0.09 |
SRRM3 |
serine/arginine repetitive matrix 3 |
15133 |
0.16 |
| chr4_1165194_1165367 | 0.09 |
SPON2 |
spondin 2, extracellular matrix protein |
1381 |
0.3 |
| chr3_192288894_192289136 | 0.09 |
ENSG00000238902 |
. |
37114 |
0.17 |
| chr3_131748397_131748548 | 0.09 |
CPNE4 |
copine IV |
5372 |
0.29 |
| chr4_7972052_7972961 | 0.09 |
AFAP1 |
actin filament associated protein 1 |
30853 |
0.14 |
| chr2_177043273_177043501 | 0.09 |
HOXD-AS1 |
HOXD cluster antisense RNA 1 |
375 |
0.73 |
| chr8_145059843_145060344 | 0.09 |
PARP10 |
poly (ADP-ribose) polymerase family, member 10 |
505 |
0.62 |
| chr11_27015640_27015894 | 0.09 |
FIBIN |
fin bud initiation factor homolog (zebrafish) |
139 |
0.98 |
| chr19_4061458_4061934 | 0.08 |
CTD-2622I13.3 |
|
1051 |
0.37 |
| chr10_79470838_79471281 | 0.08 |
ENSG00000199664 |
. |
65754 |
0.1 |
| chr1_15291094_15291245 | 0.08 |
KAZN |
kazrin, periplakin interacting protein |
18754 |
0.28 |
| chr2_946897_947048 | 0.08 |
SNTG2 |
syntrophin, gamma 2 |
347 |
0.79 |
| chr13_111524223_111524374 | 0.08 |
LINC00346 |
long intergenic non-protein coding RNA 346 |
2136 |
0.32 |
| chr5_139562716_139562882 | 0.08 |
CYSTM1 |
cysteine-rich transmembrane module containing 1 |
8572 |
0.14 |
| chr20_60980967_60981118 | 0.08 |
CABLES2 |
Cdk5 and Abl enzyme substrate 2 |
1299 |
0.34 |
| chr19_56097218_56097408 | 0.08 |
ZNF579 |
zinc finger protein 579 |
5102 |
0.09 |
| chr5_68026465_68026616 | 0.08 |
ENSG00000212249 |
. |
143241 |
0.05 |
| chr19_7927790_7927941 | 0.08 |
EVI5L |
ecotropic viral integration site 5-like |
2394 |
0.13 |
| chr18_56436184_56436335 | 0.08 |
RP11-126O1.4 |
|
16297 |
0.15 |
| chr3_48469827_48469978 | 0.08 |
PLXNB1 |
plexin B1 |
970 |
0.35 |
| chr7_73437680_73437831 | 0.08 |
ELN |
elastin |
4364 |
0.23 |
| chr19_38746743_38747049 | 0.08 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
116 |
0.93 |
| chr5_140857632_140858335 | 0.08 |
ENSG00000242020 |
. |
900 |
0.34 |
| chr9_971802_971953 | 0.08 |
DMRT3 |
doublesex and mab-3 related transcription factor 3 |
5087 |
0.26 |
| chr19_35607242_35607393 | 0.08 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
93 |
0.93 |
| chr5_135416426_135416805 | 0.08 |
ENSG00000270123 |
. |
329 |
0.89 |
| chr2_202899935_202900086 | 0.08 |
FZD7 |
frizzled family receptor 7 |
700 |
0.67 |
| chr7_15726278_15726429 | 0.08 |
MEOX2 |
mesenchyme homeobox 2 |
84 |
0.98 |
| chr5_133775184_133775365 | 0.08 |
CDKN2AIPNL |
CDKN2A interacting protein N-terminal like |
27685 |
0.14 |
| chr1_36808540_36808801 | 0.08 |
STK40 |
serine/threonine kinase 40 |
18271 |
0.12 |
| chr1_23886268_23886631 | 0.07 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
164 |
0.95 |
| chr9_136374693_136375064 | 0.07 |
TMEM8C |
transmembrane protein 8C |
15190 |
0.12 |
| chr4_121991561_121991712 | 0.07 |
NDNF |
neuron-derived neurotrophic factor |
1520 |
0.44 |
| chr11_10561561_10561712 | 0.07 |
RNF141 |
ring finger protein 141 |
817 |
0.46 |
| chr2_60759582_60759733 | 0.07 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
20883 |
0.21 |
| chr17_46674549_46675551 | 0.07 |
HOXB-AS3 |
HOXB cluster antisense RNA 3 |
1249 |
0.2 |
| chr1_236096840_236097020 | 0.07 |
ENSG00000206803 |
. |
18215 |
0.19 |
| chr6_139014672_139015008 | 0.07 |
RP11-390P2.4 |
|
616 |
0.63 |
| chr8_116674563_116674799 | 0.07 |
TRPS1 |
trichorhinophalangeal syndrome I |
776 |
0.79 |
| chr9_116937635_116937786 | 0.07 |
COL27A1 |
collagen, type XXVII, alpha 1 |
7715 |
0.22 |
| chr12_82153128_82153514 | 0.07 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
11 |
0.99 |
| chr8_37654901_37655621 | 0.07 |
GPR124 |
G protein-coupled receptor 124 |
487 |
0.76 |
| chr19_9608666_9608918 | 0.07 |
ZNF560 |
zinc finger protein 560 |
491 |
0.79 |
| chr1_32263785_32263971 | 0.07 |
SPOCD1 |
SPOC domain containing 1 |
439 |
0.79 |
| chr7_116312458_116312799 | 0.07 |
MET |
met proto-oncogene |
169 |
0.97 |
| chr4_2262463_2263167 | 0.07 |
MXD4 |
MAX dimerization protein 4 |
1206 |
0.35 |
| chr19_49220034_49220185 | 0.07 |
MAMSTR |
MEF2 activating motif and SAP domain containing transcriptional regulator |
9 |
0.95 |
| chr15_74466491_74466642 | 0.07 |
ISLR |
immunoglobulin superfamily containing leucine-rich repeat |
95 |
0.95 |
| chr16_70719186_70719575 | 0.07 |
MTSS1L |
metastasis suppressor 1-like |
589 |
0.67 |
| chr7_101376826_101377146 | 0.07 |
CUX1 |
cut-like homeobox 1 |
81973 |
0.09 |
| chr8_143463736_143463887 | 0.07 |
TSNARE1 |
t-SNARE domain containing 1 |
20438 |
0.18 |
| chr13_95655119_95655331 | 0.07 |
ENSG00000252335 |
. |
16264 |
0.29 |
| chr2_153191641_153191918 | 0.07 |
FMNL2 |
formin-like 2 |
28 |
0.99 |
| chr20_61603893_61604112 | 0.07 |
SLC17A9 |
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
19604 |
0.14 |
| chr16_79674778_79674948 | 0.07 |
ENSG00000221330 |
. |
28771 |
0.22 |
| chr7_42272520_42272675 | 0.07 |
GLI3 |
GLI family zinc finger 3 |
4015 |
0.38 |
| chr4_75174743_75174922 | 0.07 |
EPGN |
epithelial mitogen |
18 |
0.98 |
| chr1_54715687_54715880 | 0.07 |
SSBP3 |
single stranded DNA binding protein 3 |
1750 |
0.29 |
| chr20_4129930_4130735 | 0.07 |
SMOX |
spermine oxidase |
750 |
0.71 |
| chr17_71301289_71301523 | 0.07 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
6141 |
0.2 |
| chr8_124552248_124552666 | 0.07 |
FBXO32 |
F-box protein 32 |
989 |
0.58 |
| chr13_74315255_74315493 | 0.07 |
KLF12 |
Kruppel-like factor 12 |
253812 |
0.02 |
| chr1_27883616_27883836 | 0.07 |
RP1-159A19.4 |
|
31410 |
0.13 |
| chr1_147717990_147718807 | 0.07 |
ENSG00000199879 |
. |
17353 |
0.18 |
| chr18_57364455_57364606 | 0.07 |
CCBE1 |
collagen and calcium binding EGF domains 1 |
82 |
0.93 |
| chr15_63233274_63233425 | 0.07 |
RP11-1069G10.1 |
|
57116 |
0.12 |
| chr3_97540946_97541232 | 0.07 |
CRYBG3 |
Beta/gamma crystallin domain-containing protein 3; cDNA FLJ60082, weakly similar to Uro-adherence factor A |
205 |
0.95 |
| chr3_9358471_9358677 | 0.07 |
THUMPD3 |
THUMP domain containing 3 |
45952 |
0.11 |
| chr16_2070297_2070477 | 0.07 |
NPW |
neuropeptide W |
841 |
0.28 |
| chr22_36902639_36903091 | 0.06 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
204 |
0.92 |
| chr4_174449079_174449230 | 0.06 |
HAND2-AS1 |
HAND2 antisense RNA 1 (head to head) |
597 |
0.73 |
| chr12_31522871_31523058 | 0.06 |
ENSG00000207477 |
. |
3742 |
0.2 |
| chr3_133614813_133615159 | 0.06 |
RAB6B |
RAB6B, member RAS oncogene family |
306 |
0.92 |
| chr9_37904898_37905049 | 0.06 |
SLC25A51 |
solute carrier family 25, member 51 |
843 |
0.66 |
| chr20_3660344_3660495 | 0.06 |
ADAM33 |
ADAM metallopeptidase domain 33 |
2331 |
0.23 |
| chr19_32827571_32827722 | 0.06 |
ZNF507 |
zinc finger protein 507 |
8854 |
0.19 |
| chr12_2411421_2411572 | 0.06 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
32554 |
0.21 |
| chr7_97839825_97840267 | 0.06 |
BHLHA15 |
basic helix-loop-helix family, member a15 |
693 |
0.72 |
| chr12_120799158_120799309 | 0.06 |
MSI1 |
musashi RNA-binding protein 1 |
6639 |
0.13 |
| chr16_73059505_73059673 | 0.06 |
AC002044.4 |
|
11214 |
0.2 |
| chr17_59539667_59539821 | 0.06 |
RP11-15K2.2 |
|
1001 |
0.49 |
| chr9_97683470_97683621 | 0.06 |
RP11-54O15.3 |
|
13683 |
0.17 |
| chr19_31617979_31618403 | 0.06 |
AC020952.1 |
Uncharacterized protein |
22171 |
0.28 |
| chr9_114908014_114908165 | 0.06 |
SUSD1 |
sushi domain containing 1 |
3343 |
0.22 |
| chr3_15599343_15599494 | 0.06 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
36160 |
0.11 |
| chr14_103488863_103489065 | 0.06 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
34835 |
0.14 |
| chr20_38615346_38615589 | 0.06 |
ENSG00000263518 |
. |
116531 |
0.07 |
| chr19_13107353_13108160 | 0.06 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
1104 |
0.3 |
| chr19_1154555_1155129 | 0.06 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
293 |
0.85 |
| chr16_4422135_4423224 | 0.06 |
VASN |
vasorin |
830 |
0.49 |
| chr19_2056138_2056609 | 0.06 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
5130 |
0.12 |
| chr17_4981225_4982338 | 0.06 |
ZFP3 |
ZFP3 zinc finger protein |
238 |
0.6 |
| chr2_224904165_224904565 | 0.06 |
SERPINE2 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
329 |
0.92 |
| chr2_192110869_192111020 | 0.06 |
MYO1B |
myosin IB |
65 |
0.98 |
| chr9_129884150_129884930 | 0.06 |
ANGPTL2 |
angiopoietin-like 2 |
373 |
0.9 |
| chr22_19794405_19794556 | 0.06 |
C22orf29 |
chromosome 22 open reading frame 29 |
47859 |
0.1 |
| chr2_103103706_103103917 | 0.06 |
SLC9A4 |
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
14049 |
0.18 |
| chr3_138153080_138153961 | 0.06 |
ESYT3 |
extended synaptotagmin-like protein 3 |
53 |
0.98 |
| chr1_2374865_2375016 | 0.06 |
PLCH2 |
phospholipase C, eta 2 |
17521 |
0.1 |
| chr2_151332783_151332934 | 0.06 |
RND3 |
Rho family GTPase 3 |
9038 |
0.33 |
| chr11_286011_286162 | 0.06 |
RP11-326C3.2 |
|
1219 |
0.22 |
| chr10_69895923_69896335 | 0.06 |
ENSG00000222371 |
. |
21707 |
0.17 |
| chr10_100027380_100027945 | 0.06 |
LOXL4 |
lysyl oxidase-like 4 |
345 |
0.9 |
| chr8_37653908_37654059 | 0.06 |
GPR124 |
G protein-coupled receptor 124 |
441 |
0.79 |
| chr11_119211431_119211655 | 0.06 |
C1QTNF5 |
C1q and tumor necrosis factor related protein 5 |
50 |
0.94 |
| chrX_3263508_3263724 | 0.06 |
MXRA5 |
matrix-remodelling associated 5 |
1066 |
0.64 |
| chr3_98618719_98618976 | 0.06 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
1168 |
0.44 |
| chr3_100185050_100185278 | 0.06 |
TMEM45A |
transmembrane protein 45A |
26299 |
0.19 |
| chr15_75639432_75640393 | 0.06 |
NEIL1 |
nei endonuclease VIII-like 1 (E. coli) |
48 |
0.96 |
| chr5_7289635_7289990 | 0.06 |
CTD-2296D1.4 |
|
83375 |
0.1 |
| chr3_42922021_42922670 | 0.06 |
CYP8B1 |
cytochrome P450, family 8, subfamily B, polypeptide 1 |
4712 |
0.15 |
| chr20_5452381_5452567 | 0.06 |
RP5-1022P6.5 |
|
61800 |
0.13 |
| chr12_26304310_26304461 | 0.06 |
SSPN |
sarcospan |
16485 |
0.18 |
| chr2_235406000_235406197 | 0.06 |
ARL4C |
ADP-ribosylation factor-like 4C |
401 |
0.92 |
| chr6_44886900_44887051 | 0.06 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
36272 |
0.23 |
| chr1_110210893_110211366 | 0.06 |
GSTM2 |
glutathione S-transferase mu 2 (muscle) |
415 |
0.7 |
| chr6_37666994_37667145 | 0.06 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
13 |
0.99 |
| chr8_145008502_145008738 | 0.06 |
PLEC |
plectin |
5138 |
0.12 |
| chr2_43298278_43298429 | 0.06 |
ENSG00000207087 |
. |
20279 |
0.27 |
| chr11_94509773_94509974 | 0.06 |
AMOTL1 |
angiomotin like 1 |
8336 |
0.24 |
| chr8_494269_494491 | 0.06 |
TDRP |
testis development related protein |
457 |
0.88 |
| chr22_28114117_28114268 | 0.05 |
RP11-375H17.1 |
|
1724 |
0.5 |
| chr20_36531075_36531364 | 0.05 |
VSTM2L |
V-set and transmembrane domain containing 2 like |
280 |
0.93 |
| chr19_2426530_2427480 | 0.05 |
TIMM13 |
translocase of inner mitochondrial membrane 13 homolog (yeast) |
531 |
0.7 |
| chr1_12677001_12677791 | 0.05 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
341 |
0.87 |
| chr7_578206_578357 | 0.05 |
ENSG00000252986 |
. |
12089 |
0.17 |
| chr13_103381776_103381971 | 0.05 |
CCDC168 |
coiled-coil domain containing 168 |
7286 |
0.13 |
| chr14_98603340_98603491 | 0.05 |
C14orf64 |
chromosome 14 open reading frame 64 |
158954 |
0.04 |
| chr12_53625173_53625934 | 0.05 |
RARG |
retinoic acid receptor, gamma |
405 |
0.74 |
| chr2_177001412_177001990 | 0.05 |
HOXD3 |
homeobox D3 |
16 |
0.5 |
| chr3_52552947_52553103 | 0.05 |
STAB1 |
stabilin 1 |
4269 |
0.13 |
| chr16_11680564_11680715 | 0.05 |
LITAF |
lipopolysaccharide-induced TNF factor |
120 |
0.97 |
| chr1_215261434_215261585 | 0.05 |
KCNK2 |
potassium channel, subfamily K, member 2 |
4649 |
0.37 |
| chr8_144302927_144303221 | 0.05 |
GPIHBP1 |
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
8006 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0044091 | membrane biogenesis(GO:0044091) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0014808 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 | 0.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |