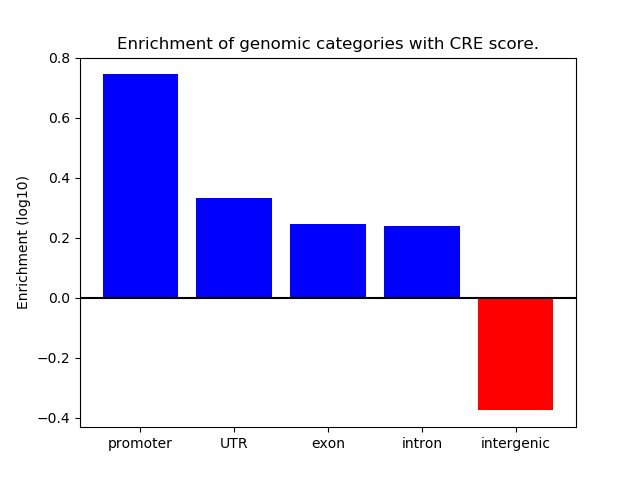
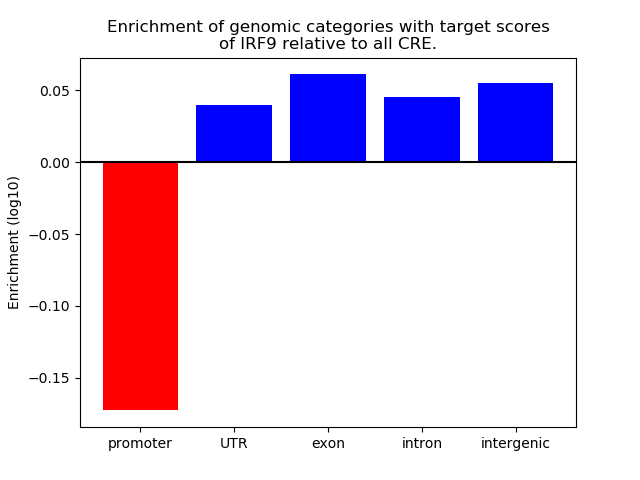
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
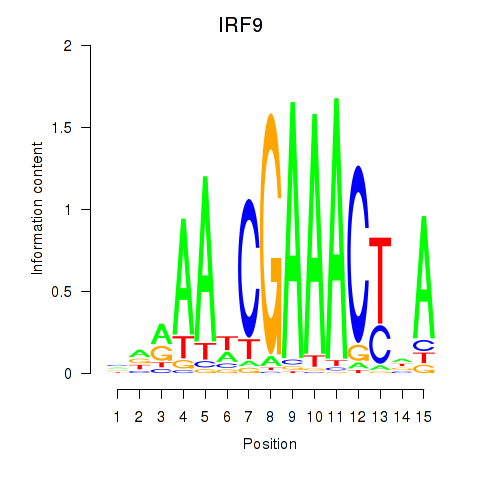
Results for IRF9
Z-value: 1.60
Transcription factors associated with IRF9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF9
|
ENSG00000213928.4 | interferon regulatory factor 9 |
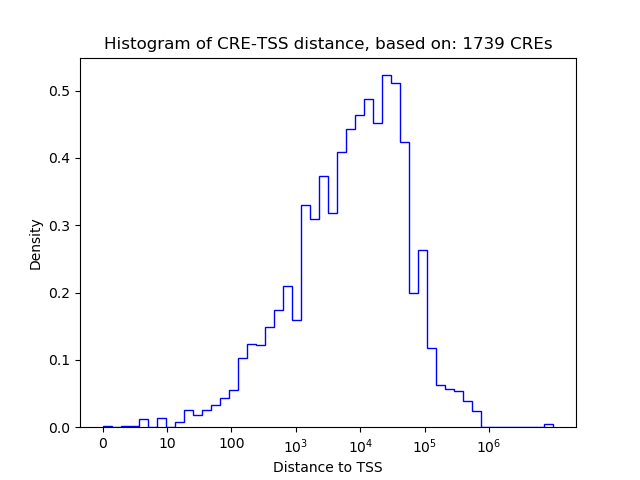
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_24632071_24632222 | IRF9 | 59 | 0.907519 | -0.54 | 1.3e-01 | Click! |
| chr14_24630498_24631577 | IRF9 | 527 | 0.470111 | 0.24 | 5.4e-01 | Click! |
| chr14_24632318_24632469 | IRF9 | 188 | 0.816439 | 0.22 | 5.6e-01 | Click! |
| chr14_24634563_24634714 | IRF9 | 2433 | 0.091041 | -0.21 | 5.8e-01 | Click! |
| chr14_24636130_24636281 | IRF9 | 4000 | 0.067793 | -0.08 | 8.4e-01 | Click! |
Activity of the IRF9 motif across conditions
Conditions sorted by the z-value of the IRF9 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
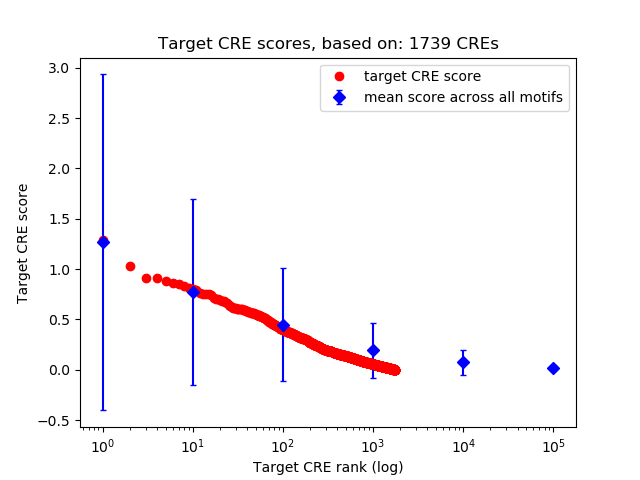
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_75263884_75264078 | 1.29 |
EREG |
epiregulin |
33121 |
0.14 |
| chr13_49832065_49832609 | 1.03 |
CDADC1 |
cytidine and dCMP deaminase domain containing 1 |
10153 |
0.23 |
| chr4_15549233_15549384 | 0.92 |
RP11-799M12.2 |
|
16568 |
0.21 |
| chr1_78392239_78392485 | 0.91 |
NEXN |
nexilin (F actin binding protein) |
8549 |
0.18 |
| chr6_44900994_44901181 | 0.89 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
22160 |
0.28 |
| chr9_139990412_139990681 | 0.86 |
ENSG00000199411 |
. |
1749 |
0.12 |
| chr7_140075788_140076381 | 0.85 |
ENSG00000238868 |
. |
618 |
0.63 |
| chr2_240302618_240303103 | 0.83 |
HDAC4 |
histone deacetylase 4 |
19783 |
0.17 |
| chr7_3134520_3134947 | 0.81 |
CARD11 |
caspase recruitment domain family, member 11 |
51154 |
0.14 |
| chr2_158301478_158301629 | 0.80 |
CYTIP |
cytohesin 1 interacting protein |
899 |
0.62 |
| chr7_2445424_2445909 | 0.79 |
CHST12 |
carbohydrate (chondroitin 4) sulfotransferase 12 |
1932 |
0.35 |
| chr16_25060772_25060995 | 0.76 |
ARHGAP17 |
Rho GTPase activating protein 17 |
33896 |
0.18 |
| chr8_117396611_117396811 | 0.76 |
ENSG00000264815 |
. |
97123 |
0.09 |
| chr2_86079968_86080395 | 0.75 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
14596 |
0.17 |
| chrX_25086022_25086209 | 0.75 |
ENSG00000240439 |
. |
7059 |
0.28 |
| chr17_6659349_6659686 | 0.74 |
XAF1 |
XIAP associated factor 1 |
131 |
0.95 |
| chr8_142244970_142245137 | 0.71 |
SLC45A4 |
solute carrier family 45, member 4 |
4606 |
0.17 |
| chr13_28696488_28696678 | 0.71 |
PAN3-AS1 |
PAN3 antisense RNA 1 |
15747 |
0.17 |
| chr12_28406339_28406704 | 0.71 |
CCDC91 |
coiled-coil domain containing 91 |
3137 |
0.33 |
| chr6_152504718_152504928 | 0.70 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
15324 |
0.29 |
| chr9_90664194_90664345 | 0.69 |
ENSG00000271923 |
. |
51019 |
0.15 |
| chr2_40008252_40008657 | 0.68 |
THUMPD2 |
THUMP domain containing 2 |
2047 |
0.37 |
| chr5_58055562_58056142 | 0.67 |
RP11-479O16.1 |
|
24754 |
0.24 |
| chr21_37670502_37670679 | 0.67 |
MORC3 |
MORC family CW-type zinc finger 3 |
21897 |
0.13 |
| chr2_162101279_162101855 | 0.65 |
AC009299.2 |
|
6106 |
0.21 |
| chr5_126098116_126098351 | 0.64 |
ENSG00000252185 |
. |
7124 |
0.23 |
| chr7_139680542_139680798 | 0.62 |
PARP12 |
poly (ADP-ribose) polymerase family, member 12 |
46448 |
0.16 |
| chr12_121476370_121477291 | 0.62 |
OASL |
2'-5'-oligoadenylate synthetase-like |
80 |
0.96 |
| chr1_45516840_45517014 | 0.62 |
ENSG00000264650 |
. |
17235 |
0.16 |
| chr11_104669483_104669764 | 0.61 |
CASP12 |
caspase 12 (gene/pseudogene) |
99518 |
0.08 |
| chr21_15914339_15914644 | 0.61 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
4171 |
0.29 |
| chr13_28365022_28365726 | 0.61 |
GSX1 |
GS homeobox 1 |
1406 |
0.47 |
| chr10_135093012_135093163 | 0.60 |
ADAM8 |
ADAM metallopeptidase domain 8 |
2715 |
0.14 |
| chr14_50366151_50366383 | 0.60 |
ENSG00000251929 |
. |
2401 |
0.2 |
| chr1_42995299_42995540 | 0.60 |
CCDC30 |
coiled-coil domain containing 30 |
5141 |
0.22 |
| chr7_7298568_7298818 | 0.60 |
C1GALT1 |
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
24742 |
0.22 |
| chrX_12986243_12986394 | 0.60 |
TMSB4X |
thymosin beta 4, X-linked |
6909 |
0.23 |
| chr21_25801249_25801648 | 0.60 |
ENSG00000232512 |
. |
671997 |
0.0 |
| chr17_29485980_29486219 | 0.58 |
RP11-142O6.1 |
|
25899 |
0.14 |
| chr22_40858602_40858925 | 0.58 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
659 |
0.7 |
| chr13_31348514_31348665 | 0.58 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
38944 |
0.19 |
| chr21_42733772_42734512 | 0.57 |
MX2 |
myxovirus (influenza virus) resistance 2 (mouse) |
208 |
0.95 |
| chr19_47026380_47026531 | 0.57 |
PNMAL2 |
paraneoplastic Ma antigen family-like 2 |
26700 |
0.11 |
| chr18_60173458_60173677 | 0.57 |
ZCCHC2 |
zinc finger, CCHC domain containing 2 |
16673 |
0.25 |
| chr10_26109879_26110030 | 0.57 |
ENSG00000251711 |
. |
1599 |
0.44 |
| chr2_240217343_240217562 | 0.56 |
ENSG00000265215 |
. |
9705 |
0.19 |
| chr3_192201108_192201259 | 0.56 |
FGF12-AS2 |
FGF12 antisense RNA 2 |
31628 |
0.16 |
| chr12_68383944_68384095 | 0.56 |
IFNG-AS1 |
IFNG antisense RNA 1 |
710 |
0.81 |
| chr7_106507255_106507406 | 0.56 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
1406 |
0.54 |
| chr15_55973905_55974056 | 0.55 |
RP11-420M1.2 |
|
1397 |
0.45 |
| chr1_85645360_85645511 | 0.55 |
SYDE2 |
synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
21294 |
0.16 |
| chr5_39201497_39202174 | 0.55 |
FYB |
FYN binding protein |
1294 |
0.59 |
| chr17_76768494_76768645 | 0.54 |
CYTH1 |
cytohesin 1 |
9785 |
0.19 |
| chr13_41068422_41068589 | 0.54 |
AL133318.1 |
Uncharacterized protein |
42818 |
0.18 |
| chr3_114913699_114913850 | 0.53 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
47656 |
0.2 |
| chr1_67235326_67235808 | 0.53 |
TCTEX1D1 |
Tctex1 domain containing 1 |
17424 |
0.16 |
| chr2_235399576_235400049 | 0.53 |
ARL4C |
ADP-ribosylation factor-like 4C |
5432 |
0.35 |
| chr6_128229281_128229524 | 0.53 |
THEMIS |
thymocyte selection associated |
7176 |
0.3 |
| chr10_15250983_15251323 | 0.53 |
RP11-25G10.2 |
|
28338 |
0.18 |
| chr4_90220151_90220996 | 0.53 |
GPRIN3 |
GPRIN family member 3 |
8588 |
0.31 |
| chr2_65047191_65047451 | 0.52 |
ENSG00000239891 |
. |
2809 |
0.25 |
| chr3_69795242_69795393 | 0.52 |
MITF |
microphthalmia-associated transcription factor |
6695 |
0.26 |
| chr5_32583768_32584065 | 0.51 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
1689 |
0.49 |
| chr5_138720842_138721347 | 0.51 |
SLC23A1 |
solute carrier family 23 (ascorbic acid transporter), member 1 |
852 |
0.41 |
| chr12_9801609_9802018 | 0.51 |
RP11-705C15.4 |
|
1208 |
0.34 |
| chr13_49079711_49079862 | 0.50 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
27427 |
0.22 |
| chr2_182029964_182030146 | 0.50 |
ENSG00000266705 |
. |
140324 |
0.05 |
| chr16_85981359_85981545 | 0.49 |
IRF8 |
interferon regulatory factor 8 |
33533 |
0.2 |
| chr8_42742851_42743002 | 0.49 |
ENSG00000242719 |
. |
6547 |
0.13 |
| chr1_8443286_8443442 | 0.49 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
18466 |
0.17 |
| chr14_91488738_91489174 | 0.48 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
37522 |
0.15 |
| chr15_65187803_65188133 | 0.47 |
ENSG00000264929 |
. |
4470 |
0.17 |
| chrX_96221078_96221229 | 0.47 |
RPA4 |
replication protein A4, 30kDa |
82246 |
0.11 |
| chr12_62658631_62659028 | 0.47 |
USP15 |
ubiquitin specific peptidase 15 |
4620 |
0.23 |
| chr2_164719052_164719232 | 0.46 |
FIGN |
fidgetin |
126620 |
0.06 |
| chrX_41190531_41191099 | 0.46 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
1836 |
0.31 |
| chr12_40077019_40077550 | 0.45 |
C12orf40 |
chromosome 12 open reading frame 40 |
57299 |
0.13 |
| chr2_26016338_26016489 | 0.45 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
13235 |
0.24 |
| chr11_82873492_82873814 | 0.45 |
PCF11 |
PCF11 cleavage and polyadenylation factor subunit |
5452 |
0.18 |
| chr11_108040783_108041103 | 0.45 |
NPAT |
nuclear protein, ataxia-telangiectasia locus |
8051 |
0.17 |
| chr7_33078848_33079779 | 0.44 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
1208 |
0.42 |
| chr14_71338943_71339141 | 0.44 |
PCNX |
pecanex homolog (Drosophila) |
35080 |
0.2 |
| chr6_13338388_13338539 | 0.44 |
TBC1D7 |
TBC1 domain family, member 7 |
9648 |
0.21 |
| chr19_35531341_35531561 | 0.44 |
HPN |
hepsin |
41 |
0.95 |
| chr2_1028039_1028190 | 0.44 |
AC114808.3 |
|
35913 |
0.18 |
| chr2_226329396_226329663 | 0.43 |
NYAP2 |
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
55932 |
0.18 |
| chr2_204718048_204718377 | 0.43 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
14297 |
0.24 |
| chr5_52336982_52337335 | 0.43 |
CTD-2366F13.2 |
|
48140 |
0.12 |
| chr5_148188804_148189074 | 0.43 |
ADRB2 |
adrenoceptor beta 2, surface |
17217 |
0.25 |
| chr1_174959503_174960178 | 0.42 |
ENSG00000252552 |
. |
5821 |
0.15 |
| chr12_93830509_93830660 | 0.42 |
UBE2N |
ubiquitin-conjugating enzyme E2N |
4448 |
0.18 |
| chr1_84609962_84610447 | 0.42 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
250 |
0.95 |
| chr14_98087128_98087308 | 0.41 |
ENSG00000240730 |
. |
90708 |
0.1 |
| chr12_10021470_10021931 | 0.41 |
CLEC2B |
C-type lectin domain family 2, member B |
1035 |
0.4 |
| chr2_175498427_175499207 | 0.41 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
490 |
0.83 |
| chr10_33775891_33776280 | 0.40 |
NRP1 |
neuropilin 1 |
150895 |
0.04 |
| chrX_119619280_119620483 | 0.40 |
LAMP2 |
lysosomal-associated membrane protein 2 |
16661 |
0.2 |
| chr1_215744331_215744482 | 0.40 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
2723 |
0.39 |
| chr17_38483011_38483310 | 0.40 |
RARA |
retinoic acid receptor, alpha |
8623 |
0.11 |
| chr1_51980736_51980982 | 0.40 |
RP11-191G24.1 |
|
3122 |
0.2 |
| chr12_66311785_66312262 | 0.40 |
AC090673.2 |
Uncharacterized protein |
5944 |
0.2 |
| chr12_27676423_27676945 | 0.40 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
320 |
0.91 |
| chr14_84755354_84755505 | 0.40 |
ENSG00000251895 |
. |
450857 |
0.01 |
| chr13_40532608_40532878 | 0.39 |
ENSG00000212553 |
. |
101379 |
0.08 |
| chr1_154915200_154915451 | 0.39 |
PMVK |
phosphomevalonate kinase |
5858 |
0.09 |
| chr16_81030456_81031156 | 0.39 |
CMC2 |
C-x(9)-C motif containing 2 |
1458 |
0.31 |
| chr17_71139370_71139564 | 0.39 |
SSTR2 |
somatostatin receptor 2 |
21684 |
0.17 |
| chr1_178045334_178045485 | 0.39 |
RASAL2 |
RAS protein activator like 2 |
17455 |
0.28 |
| chr7_154996931_154997229 | 0.38 |
AC099552.4 |
Uncharacterized protein |
6934 |
0.22 |
| chr9_139796334_139796685 | 0.38 |
TRAF2 |
TNF receptor-associated factor 2 |
12522 |
0.07 |
| chr4_154449244_154449395 | 0.38 |
KIAA0922 |
KIAA0922 |
27729 |
0.22 |
| chr4_184949529_184949975 | 0.38 |
STOX2 |
storkhead box 2 |
27274 |
0.21 |
| chr3_111043169_111043480 | 0.38 |
CD96 |
CD96 molecule |
31758 |
0.24 |
| chr1_160676943_160677601 | 0.38 |
CD48 |
CD48 molecule |
4321 |
0.18 |
| chr1_160489868_160490211 | 0.37 |
SLAMF6 |
SLAM family member 6 |
2993 |
0.18 |
| chr9_36493105_36493256 | 0.37 |
MELK |
maternal embryonic leucine zipper kinase |
79679 |
0.1 |
| chr10_70989347_70989498 | 0.37 |
RP11-227H15.4 |
|
2824 |
0.21 |
| chr13_41558712_41559488 | 0.37 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
2682 |
0.29 |
| chr3_52309275_52309582 | 0.37 |
WDR82 |
WD repeat domain 82 |
2909 |
0.12 |
| chr10_32655030_32655181 | 0.37 |
EPC1 |
enhancer of polycomb homolog 1 (Drosophila) |
12621 |
0.13 |
| chr4_82415207_82415715 | 0.37 |
RASGEF1B |
RasGEF domain family, member 1B |
22392 |
0.28 |
| chr13_74806763_74806914 | 0.36 |
ENSG00000206617 |
. |
56513 |
0.16 |
| chr3_4867321_4867754 | 0.36 |
ENSG00000239126 |
. |
52849 |
0.14 |
| chr2_201238051_201238202 | 0.36 |
ENSG00000201649 |
. |
228 |
0.93 |
| chr2_7017776_7018587 | 0.36 |
RSAD2 |
radical S-adenosyl methionine domain containing 2 |
385 |
0.85 |
| chr15_58702506_58703018 | 0.36 |
LIPC |
lipase, hepatic |
6 |
0.98 |
| chr2_38981773_38982016 | 0.36 |
GEMIN6 |
gem (nuclear organelle) associated protein 6 |
3218 |
0.21 |
| chr21_42797993_42798388 | 0.36 |
MX1 |
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
22 |
0.98 |
| chr12_51716816_51717143 | 0.35 |
BIN2 |
bridging integrator 2 |
920 |
0.52 |
| chr22_30617100_30617291 | 0.35 |
RP3-438O4.4 |
|
14097 |
0.11 |
| chr11_102187585_102188027 | 0.35 |
BIRC3 |
baculoviral IAP repeat containing 3 |
420 |
0.82 |
| chr15_57475648_57475933 | 0.35 |
TCF12 |
transcription factor 12 |
35838 |
0.21 |
| chr10_3514525_3515428 | 0.35 |
RP11-184A2.3 |
|
278283 |
0.01 |
| chr5_133407199_133407350 | 0.35 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
43128 |
0.15 |
| chr19_12807522_12807697 | 0.35 |
FBXW9 |
F-box and WD repeat domain containing 9 |
152 |
0.87 |
| chr16_68503734_68504345 | 0.35 |
ENSG00000199263 |
. |
4528 |
0.18 |
| chr7_88632213_88632768 | 0.35 |
C7orf62 |
chromosome 7 open reading frame 62 |
207455 |
0.03 |
| chr12_79364195_79364346 | 0.35 |
SYT1 |
synaptotagmin I |
6455 |
0.31 |
| chr6_159463569_159463720 | 0.34 |
TAGAP |
T-cell activation RhoGTPase activating protein |
2406 |
0.29 |
| chr4_113006479_113006630 | 0.34 |
C4orf32 |
chromosome 4 open reading frame 32 |
59999 |
0.14 |
| chr2_50167895_50168182 | 0.34 |
NRXN1 |
neurexin 1 |
33303 |
0.22 |
| chr10_31287025_31287176 | 0.34 |
ZNF438 |
zinc finger protein 438 |
1346 |
0.57 |
| chr6_25367391_25367687 | 0.33 |
ENSG00000207490 |
. |
16288 |
0.18 |
| chr5_158302177_158302618 | 0.33 |
CTD-2363C16.1 |
|
107617 |
0.07 |
| chr12_46062870_46063096 | 0.33 |
ARID2 |
AT rich interactive domain 2 (ARID, RFX-like) |
60465 |
0.15 |
| chr5_81074467_81075538 | 0.33 |
SSBP2 |
single-stranded DNA binding protein 2 |
27930 |
0.25 |
| chr12_6736731_6736882 | 0.33 |
LPAR5 |
lysophosphatidic acid receptor 5 |
4009 |
0.1 |
| chr13_48895006_48895418 | 0.33 |
RB1 |
retinoblastoma 1 |
17301 |
0.25 |
| chr14_101315771_101315992 | 0.33 |
ENSG00000214548 |
. |
236 |
0.69 |
| chr4_144326518_144326669 | 0.33 |
GAB1 |
GRB2-associated binding protein 1 |
13930 |
0.23 |
| chr17_78237440_78238417 | 0.33 |
RNF213 |
ring finger protein 213 |
3259 |
0.2 |
| chr7_144509813_144510191 | 0.32 |
TPK1 |
thiamin pyrophosphokinase 1 |
23144 |
0.23 |
| chr19_11159843_11160026 | 0.32 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
9577 |
0.14 |
| chr17_66196156_66196600 | 0.32 |
AMZ2 |
archaelysin family metallopeptidase 2 |
47337 |
0.11 |
| chr12_124941723_124942278 | 0.32 |
NCOR2 |
nuclear receptor corepressor 2 |
26315 |
0.25 |
| chr8_124049779_124050367 | 0.32 |
TBC1D31 |
TBC1 domain family, member 31 |
4135 |
0.17 |
| chr2_158295494_158296053 | 0.32 |
CYTIP |
cytohesin 1 interacting protein |
153 |
0.96 |
| chr20_49100210_49100461 | 0.32 |
PTPN1 |
protein tyrosine phosphatase, non-receptor type 1 |
26556 |
0.16 |
| chr14_90190957_90191587 | 0.32 |
ENSG00000200312 |
. |
12417 |
0.22 |
| chrY_15016828_15017148 | 0.32 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
246 |
0.96 |
| chr1_228457303_228457498 | 0.32 |
RP5-1139B12.2 |
|
4698 |
0.13 |
| chr10_70444201_70444500 | 0.32 |
RP11-119F7.5 |
|
13907 |
0.15 |
| chr3_100384136_100384305 | 0.32 |
ENSG00000252989 |
. |
19255 |
0.19 |
| chr7_30766967_30767188 | 0.32 |
INMT |
indolethylamine N-methyltransferase |
24674 |
0.16 |
| chrX_154012069_154012228 | 0.32 |
ENSG00000206693 |
. |
8875 |
0.12 |
| chrX_131624482_131624759 | 0.32 |
MBNL3 |
muscleblind-like splicing regulator 3 |
624 |
0.83 |
| chr13_26421188_26421471 | 0.31 |
AL138815.1 |
Uncharacterized protein |
20732 |
0.21 |
| chr5_158633120_158633309 | 0.31 |
RNF145 |
ring finger protein 145 |
1428 |
0.38 |
| chr10_102985313_102985579 | 0.31 |
LBX1-AS1 |
LBX1 antisense RNA 1 (head to head) |
3905 |
0.17 |
| chr5_56114546_56115097 | 0.31 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
3420 |
0.24 |
| chr3_71538892_71539043 | 0.31 |
ENSG00000221264 |
. |
52273 |
0.14 |
| chr12_15650638_15650843 | 0.31 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
48546 |
0.18 |
| chr3_153005612_153005763 | 0.31 |
ENSG00000265813 |
. |
87881 |
0.09 |
| chr6_13436227_13436378 | 0.31 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
27933 |
0.16 |
| chr3_63965361_63965766 | 0.31 |
ATXN7 |
ataxin 7 |
12143 |
0.14 |
| chr17_181340_181491 | 0.30 |
ENSG00000262061 |
. |
419 |
0.56 |
| chr10_102897939_102898090 | 0.30 |
RP11-31L23.3 |
|
1454 |
0.29 |
| chr2_181528279_181528430 | 0.30 |
ENSG00000264976 |
. |
46421 |
0.21 |
| chr5_14595155_14595651 | 0.30 |
FAM105A |
family with sequence similarity 105, member A |
13519 |
0.24 |
| chr4_90239487_90239778 | 0.30 |
GPRIN3 |
GPRIN family member 3 |
10471 |
0.3 |
| chr11_60214184_60214345 | 0.30 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
8961 |
0.16 |
| chr19_4915707_4915936 | 0.30 |
ARRDC5 |
arrestin domain containing 5 |
12942 |
0.12 |
| chr11_122748654_122748805 | 0.30 |
C11orf63 |
chromosome 11 open reading frame 63 |
4662 |
0.22 |
| chr10_173746_173998 | 0.30 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
6533 |
0.24 |
| chr12_9884469_9884620 | 0.30 |
CLECL1 |
C-type lectin-like 1 |
1171 |
0.42 |
| chr1_121200380_121200843 | 0.29 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
264674 |
0.02 |
| chr5_139047564_139047990 | 0.29 |
CXXC5 |
CXXC finger protein 5 |
7244 |
0.22 |
| chr7_7287609_7287760 | 0.29 |
C1GALT1 |
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
13733 |
0.24 |
| chr15_60942503_60942673 | 0.29 |
RORA |
RAR-related orphan receptor A |
22926 |
0.19 |
| chr5_118673119_118673270 | 0.29 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
4324 |
0.24 |
| chr3_125654138_125654289 | 0.29 |
ALG1L |
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase-like |
1669 |
0.35 |
| chrX_23791425_23791576 | 0.28 |
ACOT9 |
acyl-CoA thioesterase 9 |
6908 |
0.14 |
| chr15_68591382_68591806 | 0.28 |
FEM1B |
fem-1 homolog b (C. elegans) |
9044 |
0.21 |
| chr20_52204433_52204907 | 0.28 |
ZNF217 |
zinc finger protein 217 |
5034 |
0.21 |
| chr13_52769267_52769418 | 0.27 |
MRPS31P5 |
mitochondrial ribosomal protein S31 pseudogene 5 |
829 |
0.66 |
| chr1_62905567_62905816 | 0.27 |
USP1 |
ubiquitin specific peptidase 1 |
3365 |
0.33 |
| chr20_8194475_8194883 | 0.27 |
PLCB1-IT1 |
PLCB1 intronic transcript 1 (non-protein coding) |
34672 |
0.21 |
| chr8_102543926_102544126 | 0.27 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
39040 |
0.17 |
| chr18_77157446_77157699 | 0.27 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
1347 |
0.51 |
| chr13_24540637_24541010 | 0.27 |
SPATA13 |
spermatogenesis associated 13 |
13121 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.3 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.1 | 0.3 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 | 0.2 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.1 | 0.2 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.2 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.1 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.3 | GO:0032727 | positive regulation of interferon-alpha production(GO:0032727) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 1.9 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.2 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0002475 | antigen processing and presentation via MHC class Ib(GO:0002475) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.4 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.1 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0046719 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.0 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.0 | GO:0010560 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0010042 | response to manganese ion(GO:0010042) |
| 0.0 | 0.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.0 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.2 | GO:0071445 | obsolete cellular response to protein stimulus(GO:0071445) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.0 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0009595 | detection of biotic stimulus(GO:0009595) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.0 | 0.0 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 0.0 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 | 0.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.0 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0015919 | peroxisomal membrane transport(GO:0015919) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.6 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.0 | GO:1903514 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.0 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.3 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.0 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.4 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.3 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.2 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.0 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |