Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for IRX6_IRX4
Z-value: 1.00
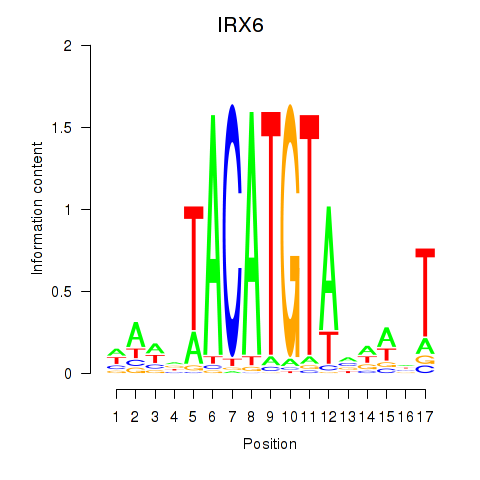
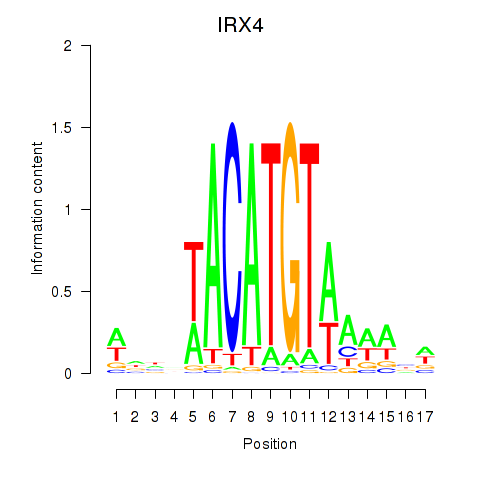
Transcription factors associated with IRX6_IRX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX6
|
ENSG00000159387.7 | iroquois homeobox 6 |
|
IRX4
|
ENSG00000113430.5 | iroquois homeobox 4 |
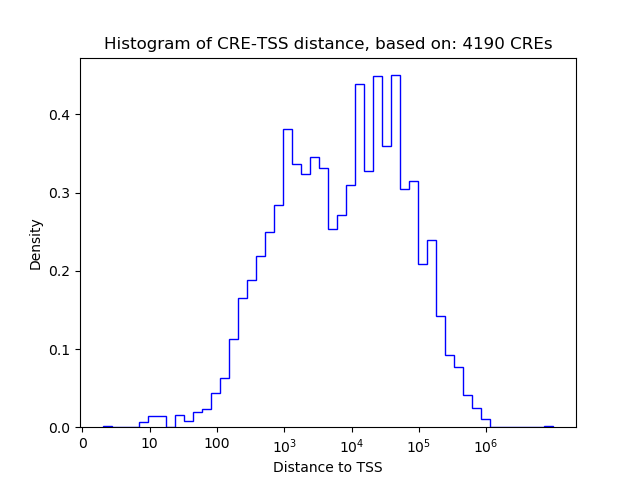
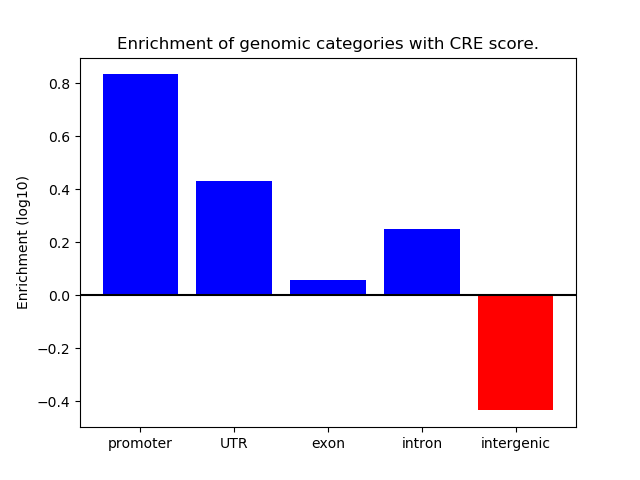
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_1887316_1887467 | IRX4 | 41 | 0.506685 | 0.56 | 1.2e-01 | Click! |
| chr5_1879697_1879902 | IRX4 | 3059 | 0.199626 | 0.53 | 1.4e-01 | Click! |
| chr5_1886065_1886216 | IRX4 | 1192 | 0.310660 | 0.44 | 2.4e-01 | Click! |
| chr5_1882927_1883078 | IRX4 | 122 | 0.926819 | 0.34 | 3.7e-01 | Click! |
| chr5_1886412_1886563 | IRX4 | 845 | 0.392112 | 0.32 | 4.0e-01 | Click! |
| chr16_55361576_55361727 | IRX6 | 3979 | 0.225858 | 0.55 | 1.2e-01 | Click! |
| chr16_55358539_55358690 | IRX6 | 942 | 0.579550 | 0.55 | 1.3e-01 | Click! |
| chr16_55359549_55359700 | IRX6 | 1952 | 0.334397 | 0.48 | 1.9e-01 | Click! |
| chr16_55358328_55358505 | IRX6 | 744 | 0.671458 | 0.26 | 4.9e-01 | Click! |
| chr16_55358802_55358953 | IRX6 | 1205 | 0.484201 | 0.26 | 5.0e-01 | Click! |
Activity of the IRX6_IRX4 motif across conditions
Conditions sorted by the z-value of the IRX6_IRX4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
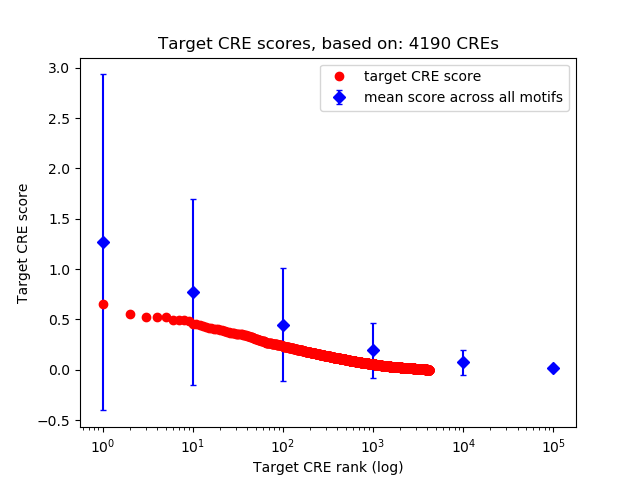
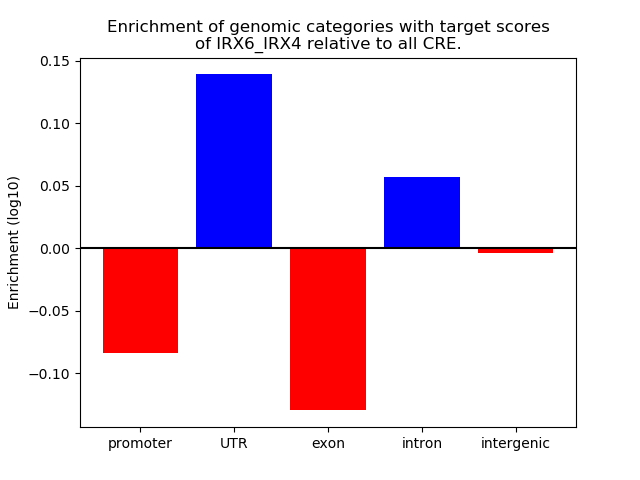
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_150779976_150780304 | 0.66 |
CTSK |
cathepsin K |
230 |
0.9 |
| chr14_81789363_81789514 | 0.56 |
STON2 |
stonin 2 |
46161 |
0.18 |
| chr2_166031255_166031406 | 0.52 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
11734 |
0.22 |
| chr6_56414295_56414544 | 0.52 |
DST |
dystonin |
75631 |
0.11 |
| chr7_117720696_117720847 | 0.52 |
NAA38 |
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
103315 |
0.08 |
| chr10_21688030_21688514 | 0.50 |
ENSG00000207264 |
. |
77382 |
0.08 |
| chr12_13351250_13351500 | 0.50 |
EMP1 |
epithelial membrane protein 1 |
1655 |
0.47 |
| chr2_173686973_173687124 | 0.49 |
RAPGEF4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
745 |
0.77 |
| chr2_189834539_189834690 | 0.49 |
COL3A1 |
collagen, type III, alpha 1 |
4432 |
0.25 |
| chr20_15680912_15681288 | 0.46 |
ENSG00000212111 |
. |
141987 |
0.05 |
| chr10_52687623_52687774 | 0.45 |
A1CF |
APOBEC1 complementation factor |
42263 |
0.16 |
| chr10_29879098_29879625 | 0.44 |
ENSG00000216035 |
. |
11914 |
0.18 |
| chr3_99360396_99360669 | 0.43 |
COL8A1 |
collagen, type VIII, alpha 1 |
3078 |
0.35 |
| chr12_42982187_42982354 | 0.43 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
1208 |
0.6 |
| chr4_187491209_187491867 | 0.42 |
MTNR1A |
melatonin receptor 1A |
14817 |
0.17 |
| chr3_71295420_71295571 | 0.41 |
FOXP1 |
forkhead box P1 |
1179 |
0.61 |
| chr3_63748246_63748457 | 0.40 |
AC136289.1 |
|
42418 |
0.14 |
| chr8_30278844_30278995 | 0.40 |
RBPMS |
RNA binding protein with multiple splicing |
21214 |
0.21 |
| chr1_197167655_197168814 | 0.40 |
ZBTB41 |
zinc finger and BTB domain containing 41 |
1438 |
0.4 |
| chr1_57661483_57661634 | 0.40 |
DAB1 |
Dab, reelin signal transducer, homolog 1 (Drosophila) |
95179 |
0.09 |
| chr11_121969171_121969347 | 0.40 |
ENSG00000207971 |
. |
1293 |
0.36 |
| chr4_170189564_170189726 | 0.39 |
SH3RF1 |
SH3 domain containing ring finger 1 |
1463 |
0.54 |
| chr19_16179581_16179939 | 0.38 |
TPM4 |
tropomyosin 4 |
1250 |
0.44 |
| chr7_42228970_42229243 | 0.38 |
GLI3 |
GLI family zinc finger 3 |
38214 |
0.24 |
| chr6_80248235_80248430 | 0.38 |
LCA5 |
Leber congenital amaurosis 5 |
1157 |
0.64 |
| chr7_116483846_116484032 | 0.37 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
18588 |
0.19 |
| chr1_69950952_69951103 | 0.36 |
LRRC7 |
leucine rich repeat containing 7 |
83054 |
0.11 |
| chr1_24739398_24739744 | 0.36 |
STPG1 |
sperm-tail PG-rich repeat containing 1 |
644 |
0.68 |
| chr3_98617426_98617589 | 0.36 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
2508 |
0.26 |
| chr9_139684873_139685024 | 0.36 |
TMEM141 |
transmembrane protein 141 |
859 |
0.25 |
| chr2_64017838_64018067 | 0.36 |
UGP2 |
UDP-glucose pyrophosphorylase 2 |
50122 |
0.16 |
| chr11_8830627_8830889 | 0.35 |
ST5 |
suppression of tumorigenicity 5 |
1438 |
0.35 |
| chr2_9398911_9399239 | 0.35 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
52181 |
0.16 |
| chr5_16549957_16550209 | 0.35 |
FAM134B |
family with sequence similarity 134, member B |
40976 |
0.15 |
| chr9_110247534_110247685 | 0.35 |
KLF4 |
Kruppel-like factor 4 (gut) |
3802 |
0.26 |
| chr6_73262911_73263125 | 0.35 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
68502 |
0.12 |
| chr4_177701020_177701171 | 0.34 |
VEGFC |
vascular endothelial growth factor C |
12786 |
0.3 |
| chr16_49686685_49686863 | 0.34 |
ZNF423 |
zinc finger protein 423 |
11362 |
0.24 |
| chr2_178196983_178197134 | 0.34 |
AC074286.1 |
|
9166 |
0.12 |
| chr12_76271592_76272084 | 0.33 |
ENSG00000243420 |
. |
80072 |
0.1 |
| chr14_64389363_64389514 | 0.33 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
13622 |
0.25 |
| chr2_120064088_120064263 | 0.33 |
STEAP3-AS1 |
STEAP3 antisense RNA 1 |
57528 |
0.1 |
| chr15_71035707_71036127 | 0.33 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
19852 |
0.21 |
| chr1_192775406_192775557 | 0.33 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
2690 |
0.37 |
| chr4_47488156_47488587 | 0.32 |
ATP10D |
ATPase, class V, type 10D |
1055 |
0.56 |
| chr1_33497639_33497820 | 0.32 |
AK2 |
adenylate kinase 2 |
4712 |
0.19 |
| chr2_10036128_10036279 | 0.32 |
TAF1B |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa |
20076 |
0.2 |
| chr4_100739524_100739675 | 0.32 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
1596 |
0.47 |
| chr18_21264983_21265134 | 0.31 |
LAMA3 |
laminin, alpha 3 |
4349 |
0.25 |
| chr18_67960362_67960513 | 0.31 |
SOCS6 |
suppressor of cytokine signaling 6 |
3398 |
0.3 |
| chr2_40078902_40079211 | 0.31 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
65463 |
0.13 |
| chr12_96769647_96769798 | 0.30 |
CDK17 |
cyclin-dependent kinase 17 |
23420 |
0.18 |
| chr2_145276293_145276624 | 0.30 |
ZEB2-AS1 |
ZEB2 antisense RNA 1 |
434 |
0.52 |
| chr6_41042761_41042912 | 0.30 |
NFYA |
nuclear transcription factor Y, alpha |
2114 |
0.2 |
| chr2_182758331_182758678 | 0.30 |
SSFA2 |
sperm specific antigen 2 |
1581 |
0.37 |
| chr1_200375128_200376098 | 0.29 |
ZNF281 |
zinc finger protein 281 |
3507 |
0.37 |
| chr19_30436184_30436558 | 0.29 |
URI1 |
URI1, prefoldin-like chaperone |
2946 |
0.39 |
| chr6_144357186_144358268 | 0.29 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
28008 |
0.21 |
| chr4_114214615_114214802 | 0.29 |
ANK2 |
ankyrin 2, neuronal |
570 |
0.85 |
| chr7_120632569_120632731 | 0.29 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
2974 |
0.27 |
| chr4_102542477_102542841 | 0.28 |
ENSG00000199529 |
. |
102374 |
0.08 |
| chr15_23033242_23034084 | 0.28 |
NIPA2 |
non imprinted in Prader-Willi/Angelman syndrome 2 |
707 |
0.67 |
| chrX_124279731_124279882 | 0.28 |
ENSG00000263886 |
. |
141819 |
0.05 |
| chr4_47489411_47489562 | 0.28 |
ATP10D |
ATPase, class V, type 10D |
2170 |
0.34 |
| chr8_54624701_54625055 | 0.27 |
ATP6V1H |
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
127643 |
0.05 |
| chr11_84633973_84634163 | 0.27 |
DLG2 |
discs, large homolog 2 (Drosophila) |
156 |
0.97 |
| chr19_6103077_6103270 | 0.27 |
CTB-66B24.1 |
|
6160 |
0.15 |
| chr19_54974999_54975150 | 0.27 |
LENG9 |
leukocyte receptor cluster (LRC) member 9 |
180 |
0.88 |
| chr2_102574409_102574560 | 0.27 |
IL1R2 |
interleukin 1 receptor, type II |
33822 |
0.19 |
| chr15_73302646_73302797 | 0.27 |
NEO1 |
neogenin 1 |
41330 |
0.2 |
| chr2_223724916_223725067 | 0.27 |
ACSL3 |
acyl-CoA synthetase long-chain family member 3 |
661 |
0.77 |
| chr4_99444037_99444307 | 0.27 |
RP11-724M22.1 |
|
26657 |
0.24 |
| chr13_25034007_25034158 | 0.26 |
TPTE2P6 |
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 pseudogene 6 |
51787 |
0.12 |
| chr3_25471806_25472309 | 0.26 |
RARB |
retinoic acid receptor, beta |
2255 |
0.41 |
| chr7_7196815_7197697 | 0.26 |
C1GALT1 |
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
691 |
0.7 |
| chr11_130305065_130305271 | 0.26 |
ADAMTS8 |
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
6280 |
0.24 |
| chr3_119808572_119808744 | 0.26 |
GSK3B |
glycogen synthase kinase 3 beta |
3855 |
0.24 |
| chrX_46442901_46443074 | 0.26 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
9768 |
0.23 |
| chr15_36231470_36231621 | 0.26 |
ENSG00000265098 |
. |
12488 |
0.32 |
| chr5_66311217_66311678 | 0.26 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
10964 |
0.24 |
| chr12_76424698_76425269 | 0.26 |
PHLDA1 |
pleckstrin homology-like domain, family A, member 1 |
401 |
0.66 |
| chr7_38923632_38923783 | 0.26 |
VPS41 |
vacuolar protein sorting 41 homolog (S. cerevisiae) |
25067 |
0.24 |
| chr20_12989594_12990369 | 0.25 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
16 |
0.99 |
| chr20_42377284_42377435 | 0.25 |
GTSF1L |
gametocyte specific factor 1-like |
21721 |
0.2 |
| chr1_186156970_186157465 | 0.25 |
GS1-174L6.4 |
|
10783 |
0.22 |
| chr11_125497097_125497248 | 0.25 |
CHEK1 |
checkpoint kinase 1 |
529 |
0.7 |
| chr6_132271007_132271170 | 0.25 |
RP11-69I8.3 |
|
998 |
0.48 |
| chr13_45620875_45621062 | 0.25 |
ENSG00000223341 |
. |
13896 |
0.16 |
| chr2_189840264_189840425 | 0.25 |
COL3A1 |
collagen, type III, alpha 1 |
1245 |
0.47 |
| chr7_45958576_45958727 | 0.25 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
1626 |
0.39 |
| chr1_163039219_163039545 | 0.25 |
RGS4 |
regulator of G-protein signaling 4 |
231 |
0.96 |
| chr1_198602543_198602694 | 0.25 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
5183 |
0.27 |
| chr3_171475362_171475648 | 0.25 |
PLD1 |
phospholipase D1, phosphatidylcholine-specific |
13644 |
0.22 |
| chr19_3098811_3099227 | 0.25 |
GNA11 |
guanine nucleotide binding protein (G protein), alpha 11 (Gq class) |
4611 |
0.13 |
| chr5_77797581_77797732 | 0.24 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
47318 |
0.18 |
| chr11_114166754_114166905 | 0.24 |
NNMT |
nicotinamide N-methyltransferase |
282 |
0.93 |
| chr6_79878048_79878303 | 0.24 |
HMGN3-AS1 |
HMGN3 antisense RNA 1 |
65260 |
0.11 |
| chr4_170190845_170191352 | 0.24 |
SH3RF1 |
SH3 domain containing ring finger 1 |
10 |
0.99 |
| chr15_83951840_83952048 | 0.24 |
BNC1 |
basonuclin 1 |
127 |
0.97 |
| chr17_16931289_16931440 | 0.23 |
MPRIP |
myosin phosphatase Rho interacting protein |
14495 |
0.18 |
| chr9_92151090_92151365 | 0.23 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
38182 |
0.17 |
| chr8_104312759_104312910 | 0.23 |
FZD6 |
frizzled family receptor 6 |
1730 |
0.28 |
| chr12_115130761_115130970 | 0.23 |
TBX3 |
T-box 3 |
8896 |
0.23 |
| chr14_56666447_56666727 | 0.23 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
80760 |
0.1 |
| chr11_130030115_130030717 | 0.23 |
ST14 |
suppression of tumorigenicity 14 (colon carcinoma) |
959 |
0.66 |
| chr2_65614657_65614911 | 0.23 |
AC012370.2 |
|
6965 |
0.23 |
| chr7_38576145_38576296 | 0.23 |
ENSG00000241756 |
. |
5281 |
0.31 |
| chr9_75767359_75768171 | 0.23 |
ANXA1 |
annexin A1 |
947 |
0.7 |
| chr3_168865162_168865471 | 0.23 |
MECOM |
MDS1 and EVI1 complex locus |
206 |
0.97 |
| chr2_130515835_130516332 | 0.23 |
AC079776.2 |
|
167215 |
0.03 |
| chr3_172280513_172281223 | 0.23 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
39571 |
0.16 |
| chr5_140938191_140938342 | 0.23 |
CTD-2024I7.13 |
|
388 |
0.78 |
| chr12_22807162_22807313 | 0.23 |
ETNK1 |
ethanolamine kinase 1 |
28946 |
0.2 |
| chr3_45052751_45053653 | 0.23 |
CLEC3B |
C-type lectin domain family 3, member B |
14473 |
0.15 |
| chr12_124453376_124453888 | 0.23 |
RP11-214K3.20 |
|
506 |
0.49 |
| chr7_7736253_7736544 | 0.22 |
RPA3 |
replication protein A3, 14kDa |
21840 |
0.23 |
| chr1_109933756_109933959 | 0.22 |
SORT1 |
sortilin 1 |
2122 |
0.27 |
| chr12_42630813_42630964 | 0.22 |
YAF2 |
YY1 associated factor 2 |
714 |
0.52 |
| chr12_62381215_62381366 | 0.22 |
FAM19A2 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
117419 |
0.06 |
| chrX_133682102_133682606 | 0.22 |
ENSG00000223749 |
. |
1613 |
0.22 |
| chr9_91924793_91925270 | 0.22 |
CKS2 |
CDC28 protein kinase regulatory subunit 2 |
1082 |
0.45 |
| chr7_41426318_41426600 | 0.22 |
INHBA-AS1 |
INHBA antisense RNA 1 |
307055 |
0.01 |
| chr2_107082072_107082277 | 0.22 |
RGPD3 |
RANBP2-like and GRIP domain containing 3 |
2652 |
0.28 |
| chr3_133967997_133968801 | 0.22 |
RYK |
receptor-like tyrosine kinase |
1038 |
0.66 |
| chr3_149301834_149301985 | 0.21 |
WWTR1 |
WW domain containing transcription regulator 1 |
7870 |
0.21 |
| chr6_141587373_141587724 | 0.21 |
ENSG00000222764 |
. |
220001 |
0.02 |
| chr3_194205911_194206797 | 0.21 |
ATP13A3 |
ATPase type 13A3 |
1034 |
0.42 |
| chr18_65929417_65929568 | 0.21 |
TMX3 |
thioredoxin-related transmembrane protein 3 |
452802 |
0.01 |
| chr17_7491050_7491201 | 0.21 |
SOX15 |
SRY (sex determining region Y)-box 15 |
2265 |
0.08 |
| chr8_116670939_116671090 | 0.21 |
TRPS1 |
trichorhinophalangeal syndrome I |
2891 |
0.42 |
| chr1_100817332_100817558 | 0.21 |
CDC14A |
cell division cycle 14A |
155 |
0.96 |
| chr8_28861777_28861928 | 0.21 |
HMBOX1 |
homeobox containing 1 |
40472 |
0.11 |
| chr2_216691555_216691736 | 0.21 |
ENSG00000212055 |
. |
51997 |
0.17 |
| chr16_68123462_68123613 | 0.21 |
ENSG00000201850 |
. |
91 |
0.94 |
| chr15_92415930_92416081 | 0.21 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
18654 |
0.24 |
| chr7_80547175_80548098 | 0.21 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
863 |
0.76 |
| chr6_2377522_2377693 | 0.21 |
ENSG00000266252 |
. |
32262 |
0.24 |
| chr11_61890885_61891204 | 0.21 |
INCENP |
inner centromere protein antigens 135/155kDa |
401 |
0.85 |
| chr5_130175429_130175580 | 0.21 |
HINT1 |
histidine triad nucleotide binding protein 1 |
322816 |
0.01 |
| chr18_73180620_73180771 | 0.21 |
SMIM21 |
small integral membrane protein 21 |
41037 |
0.19 |
| chr6_42537305_42537764 | 0.21 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
5734 |
0.24 |
| chr10_10953235_10953446 | 0.20 |
CELF2 |
CUGBP, Elav-like family member 2 |
93919 |
0.08 |
| chr6_138975748_138976299 | 0.20 |
RP11-390P2.4 |
|
37662 |
0.17 |
| chr8_16689434_16690090 | 0.20 |
ENSG00000264092 |
. |
31243 |
0.24 |
| chr2_178029566_178030097 | 0.20 |
ENSG00000251746 |
. |
26404 |
0.12 |
| chr4_83810146_83810528 | 0.20 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
1834 |
0.28 |
| chr15_59939623_59939774 | 0.20 |
GTF2A2 |
general transcription factor IIA, 2, 12kDa |
9980 |
0.14 |
| chr5_12989682_12989833 | 0.20 |
ENSG00000222717 |
. |
185203 |
0.03 |
| chr1_72587992_72588143 | 0.20 |
NEGR1 |
neuronal growth regulator 1 |
21454 |
0.29 |
| chr2_147579520_147579813 | 0.20 |
ENSG00000238860 |
. |
501873 |
0.0 |
| chr5_126182453_126182725 | 0.20 |
LMNB1 |
lamin B1 |
69708 |
0.11 |
| chr10_1100415_1100678 | 0.20 |
WDR37 |
WD repeat domain 37 |
1798 |
0.26 |
| chr13_80059310_80059736 | 0.20 |
NDFIP2 |
Nedd4 family interacting protein 2 |
3935 |
0.24 |
| chr13_75466251_75466402 | 0.20 |
ENSG00000206812 |
. |
217398 |
0.02 |
| chr16_10646719_10646878 | 0.20 |
EMP2 |
epithelial membrane protein 2 |
6213 |
0.18 |
| chr6_21506218_21506369 | 0.20 |
SOX4 |
SRY (sex determining region Y)-box 4 |
87679 |
0.11 |
| chr20_35830256_35830797 | 0.20 |
MROH8 |
maestro heat-like repeat family member 8 |
22535 |
0.17 |
| chr20_52407623_52407774 | 0.19 |
ENSG00000238468 |
. |
122401 |
0.05 |
| chr3_184036946_184037180 | 0.19 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
76 |
0.94 |
| chr14_71838887_71839322 | 0.19 |
ENSG00000207444 |
. |
25950 |
0.19 |
| chr2_160569559_160570270 | 0.19 |
MARCH7 |
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
896 |
0.68 |
| chr5_121424700_121424851 | 0.19 |
LOX |
lysyl oxidase |
10795 |
0.21 |
| chr15_57511580_57511963 | 0.19 |
TCF12 |
transcription factor 12 |
107 |
0.98 |
| chr2_232646427_232646801 | 0.19 |
COPS7B |
COP9 signalosome subunit 7B |
219 |
0.79 |
| chr2_187354223_187354374 | 0.19 |
ZC3H15 |
zinc finger CCCH-type containing 15 |
3318 |
0.24 |
| chr6_47381977_47382964 | 0.19 |
ENSG00000266330 |
. |
48182 |
0.14 |
| chr3_139175617_139175768 | 0.19 |
RP11-319G6.3 |
|
9580 |
0.15 |
| chr5_53303962_53304113 | 0.19 |
ENSG00000241230 |
. |
40101 |
0.16 |
| chr9_12718902_12719101 | 0.19 |
TYRP1 |
tyrosinase-related protein 1 |
23281 |
0.2 |
| chr4_114880734_114880959 | 0.19 |
RP11-26P13.2 |
|
16434 |
0.24 |
| chr7_116229019_116229170 | 0.19 |
AC006159.4 |
|
17806 |
0.18 |
| chr21_33613293_33613449 | 0.19 |
MIS18A-AS1 |
MIS18A antisense RNA 1 |
36803 |
0.12 |
| chr8_130742298_130742449 | 0.19 |
GSDMC |
gasdermin C |
56761 |
0.14 |
| chr6_121756849_121757212 | 0.19 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
192 |
0.94 |
| chr9_92724401_92724683 | 0.19 |
ENSG00000263967 |
. |
61275 |
0.16 |
| chr16_79758893_79759044 | 0.19 |
ENSG00000221330 |
. |
55334 |
0.17 |
| chr3_42888617_42888768 | 0.18 |
ACKR2 |
atypical chemokine receptor 2 |
3236 |
0.16 |
| chr2_85778069_85778220 | 0.18 |
GGCX |
gamma-glutamyl carboxylase |
10488 |
0.09 |
| chr7_23518547_23518698 | 0.18 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
8536 |
0.17 |
| chr4_26347600_26347761 | 0.18 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
2918 |
0.4 |
| chr19_12061488_12061639 | 0.18 |
ZNF700 |
zinc finger protein 700 |
3994 |
0.13 |
| chr10_124066979_124067204 | 0.18 |
BTBD16 |
BTB (POZ) domain containing 16 |
36270 |
0.14 |
| chr4_148009310_148009488 | 0.18 |
TTC29 |
tetratricopeptide repeat domain 29 |
142365 |
0.05 |
| chr1_32168571_32169014 | 0.18 |
COL16A1 |
collagen, type XVI, alpha 1 |
976 |
0.48 |
| chr12_5540873_5541141 | 0.18 |
NTF3 |
neurotrophin 3 |
272 |
0.96 |
| chr2_145269652_145269839 | 0.18 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
5370 |
0.26 |
| chr19_4402073_4402293 | 0.18 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
476 |
0.58 |
| chr12_92541063_92541214 | 0.18 |
RP11-796E2.4 |
|
1181 |
0.36 |
| chr2_150782681_150782948 | 0.18 |
ENSG00000207270 |
. |
315526 |
0.01 |
| chr22_30485175_30485353 | 0.18 |
HORMAD2 |
HORMA domain containing 2 |
8264 |
0.18 |
| chr12_104226819_104227112 | 0.18 |
NT5DC3 |
5'-nucleotidase domain containing 3 |
8010 |
0.15 |
| chr21_16431010_16431483 | 0.18 |
NRIP1 |
nuclear receptor interacting protein 1 |
5880 |
0.3 |
| chr1_226791109_226791285 | 0.18 |
C1orf95 |
chromosome 1 open reading frame 95 |
54696 |
0.12 |
| chr2_155795915_155796066 | 0.18 |
ENSG00000223197 |
. |
150060 |
0.04 |
| chrX_130917856_130918233 | 0.18 |
ENSG00000200587 |
. |
154940 |
0.04 |
| chr10_13491532_13491683 | 0.18 |
BEND7 |
BEN domain containing 7 |
31466 |
0.14 |
| chr3_170893303_170893462 | 0.18 |
TNIK |
TRAF2 and NCK interacting kinase |
50118 |
0.15 |
| chr1_224373491_224373642 | 0.18 |
DEGS1 |
delta(4)-desaturase, sphingolipid 1 |
2598 |
0.21 |
| chr4_186696107_186696537 | 0.17 |
SORBS2 |
sorbin and SH3 domain containing 2 |
108 |
0.98 |
| chr9_112814566_112814717 | 0.17 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
3665 |
0.35 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.3 | GO:0043206 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.3 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.1 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.2 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.0 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:1903020 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.0 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.0 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.0 | GO:0016093 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.0 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031211 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME CELL CELL COMMUNICATION | Genes involved in Cell-Cell communication |