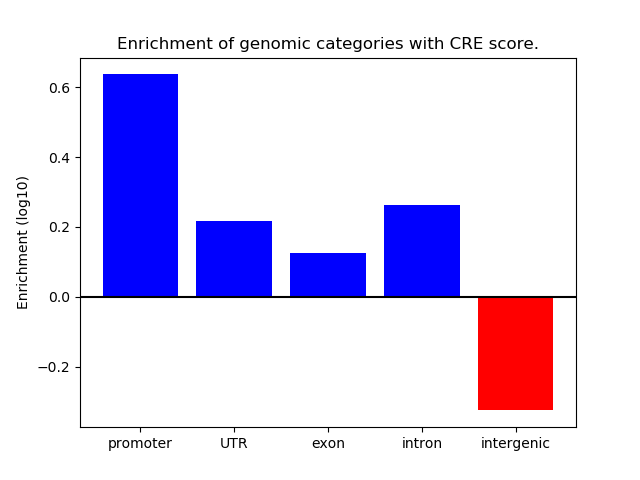
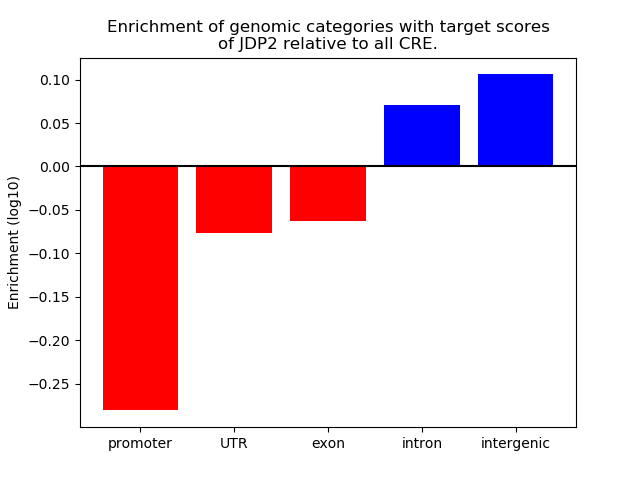
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
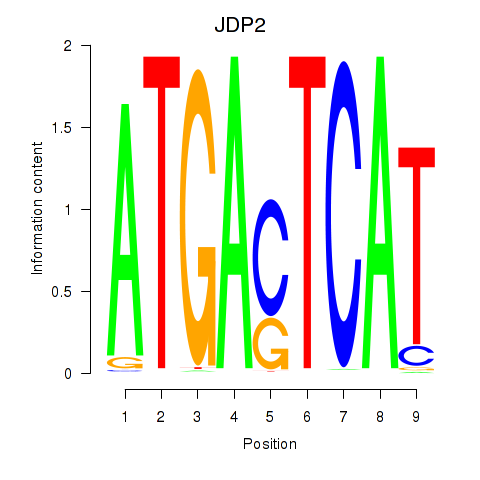
Results for JDP2
Z-value: 1.36
Transcription factors associated with JDP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JDP2
|
ENSG00000140044.8 | Jun dimerization protein 2 |
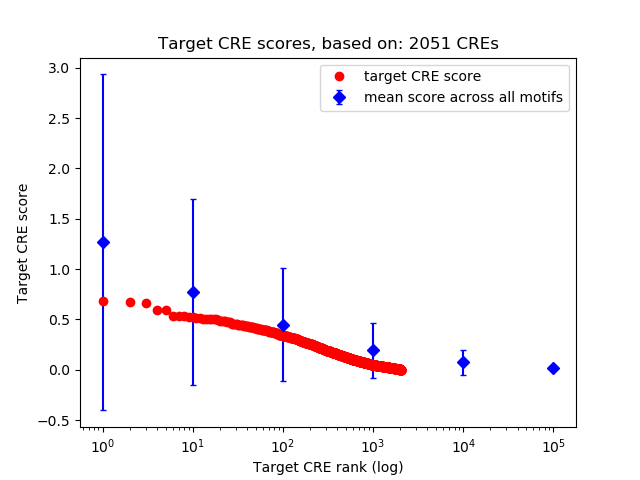
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_75925878_75926029 | JDP2 | 27116 | 0.157778 | -0.75 | 1.9e-02 | Click! |
| chr14_75925653_75925804 | JDP2 | 26891 | 0.158277 | -0.71 | 3.2e-02 | Click! |
| chr14_75920844_75920995 | JDP2 | 22082 | 0.168556 | -0.65 | 5.8e-02 | Click! |
| chr14_75919897_75920096 | JDP2 | 21159 | 0.170434 | 0.49 | 1.8e-01 | Click! |
| chr14_75923265_75923416 | JDP2 | 24503 | 0.163477 | -0.44 | 2.4e-01 | Click! |
Activity of the JDP2 motif across conditions
Conditions sorted by the z-value of the JDP2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
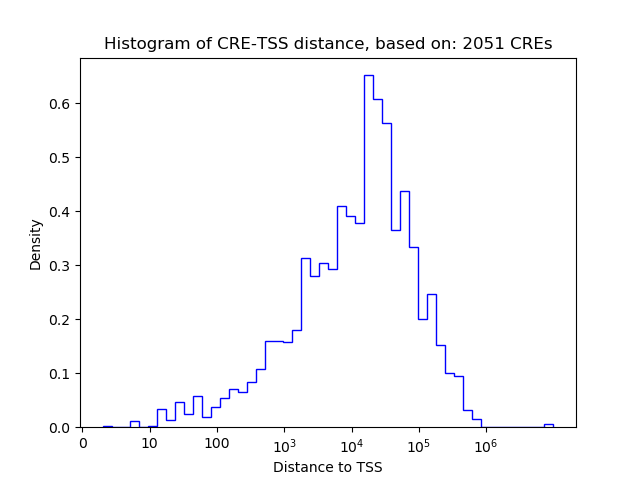
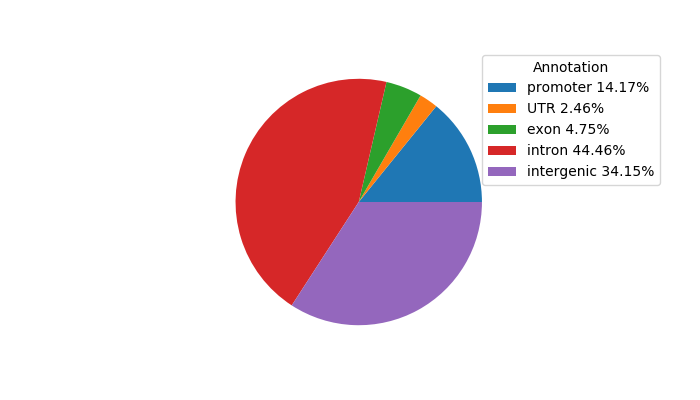
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_80603632_80603783 | 0.69 |
RP11-525K10.3 |
|
6675 |
0.18 |
| chr15_75943747_75944181 | 0.67 |
SNX33 |
sorting nexin 33 |
1867 |
0.18 |
| chr3_93878024_93878443 | 0.67 |
ENSG00000253062 |
. |
60823 |
0.12 |
| chr12_113859774_113860410 | 0.59 |
SDSL |
serine dehydratase-like |
50 |
0.97 |
| chr18_29929931_29930163 | 0.59 |
RP11-344B2.2 |
|
62647 |
0.1 |
| chr6_82854353_82854866 | 0.54 |
ENSG00000223044 |
. |
65548 |
0.14 |
| chr9_124991302_124991453 | 0.54 |
LHX6 |
LIM homeobox 6 |
157 |
0.94 |
| chr17_4762471_4762622 | 0.53 |
ENSG00000263599 |
. |
16375 |
0.08 |
| chr12_3277666_3277860 | 0.53 |
TSPAN9-IT1 |
TSPAN9 intronic transcript 1 (non-protein coding) |
18800 |
0.21 |
| chr6_44424385_44424803 | 0.52 |
ENSG00000266619 |
. |
21216 |
0.17 |
| chr13_59357512_59357663 | 0.52 |
ENSG00000222733 |
. |
255702 |
0.02 |
| chr4_158941489_158941966 | 0.51 |
FAM198B |
family with sequence similarity 198, member B |
139106 |
0.05 |
| chr1_85763681_85764097 | 0.51 |
ENSG00000264380 |
. |
13596 |
0.14 |
| chrX_48917537_48917688 | 0.51 |
CCDC120 |
coiled-coil domain containing 120 |
1066 |
0.3 |
| chr4_183047320_183047598 | 0.51 |
AC108142.1 |
|
17470 |
0.17 |
| chr1_214649424_214649729 | 0.50 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
11430 |
0.29 |
| chr11_11589938_11590089 | 0.50 |
RP11-483L5.1 |
|
1618 |
0.47 |
| chr6_72595878_72596126 | 0.50 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
404 |
0.92 |
| chr6_14001615_14001982 | 0.50 |
RNF182 |
ring finger protein 182 |
76374 |
0.11 |
| chr9_86916681_86916945 | 0.49 |
RP11-380F14.2 |
|
23679 |
0.22 |
| chr5_14268280_14268580 | 0.48 |
TRIO |
trio Rho guanine nucleotide exchange factor |
22656 |
0.28 |
| chr5_42772787_42772984 | 0.48 |
CCDC152 |
coiled-coil domain containing 152 |
15965 |
0.2 |
| chr7_110026237_110026388 | 0.48 |
AC073326.3 |
|
339569 |
0.01 |
| chr11_62308687_62309106 | 0.48 |
RP11-864I4.3 |
|
4112 |
0.1 |
| chr11_128778623_128778851 | 0.48 |
KCNJ5 |
potassium inwardly-rectifying channel, subfamily J, member 5 |
2342 |
0.24 |
| chr9_100465936_100466445 | 0.47 |
ENSG00000266608 |
. |
3674 |
0.21 |
| chr15_71126267_71126418 | 0.46 |
RP11-138H8.6 |
|
15767 |
0.15 |
| chr6_150346440_150346821 | 0.46 |
RAET1L |
retinoic acid early transcript 1L |
23 |
0.97 |
| chr2_27502961_27503177 | 0.45 |
TRIM54 |
tripartite motif containing 54 |
2191 |
0.15 |
| chr9_122573363_122573648 | 0.45 |
RP11-295D22.1 |
Uncharacterized protein |
315852 |
0.01 |
| chr1_112135126_112135664 | 0.45 |
RAP1A |
RAP1A, member of RAS oncogene family |
27004 |
0.14 |
| chr16_3692877_3693028 | 0.45 |
DNASE1 |
deoxyribonuclease I |
13 |
0.97 |
| chr8_74473490_74473704 | 0.45 |
STAU2 |
staufen double-stranded RNA binding protein 2 |
21468 |
0.27 |
| chr17_3694838_3694989 | 0.44 |
ITGAE |
integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) |
9624 |
0.15 |
| chr8_24867437_24867792 | 0.44 |
CTD-2168K21.2 |
|
53479 |
0.14 |
| chr2_91845042_91845193 | 0.44 |
AC027612.6 |
|
1588 |
0.43 |
| chr8_105677603_105677754 | 0.44 |
RP11-127H5.1 |
Uncharacterized protein |
74717 |
0.1 |
| chr11_12065337_12065964 | 0.44 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
34334 |
0.19 |
| chr1_86316102_86316253 | 0.43 |
ENSG00000201620 |
. |
33304 |
0.23 |
| chr1_221209086_221209419 | 0.43 |
HLX |
H2.0-like homeobox |
154668 |
0.04 |
| chr7_24988449_24988600 | 0.43 |
OSBPL3 |
oxysterol binding protein-like 3 |
30812 |
0.21 |
| chr19_48793598_48793806 | 0.43 |
ZNF114 |
zinc finger protein 114 |
18822 |
0.1 |
| chr4_139146799_139146992 | 0.43 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
16608 |
0.27 |
| chr1_115732540_115732691 | 0.43 |
RP4-663N10.1 |
|
93040 |
0.08 |
| chr2_100635044_100635528 | 0.43 |
AFF3 |
AF4/FMR2 family, member 3 |
85706 |
0.09 |
| chr16_12121143_12121294 | 0.42 |
SNX29 |
sorting nexin 29 |
24837 |
0.12 |
| chr19_49071767_49072143 | 0.42 |
SULT2B1 |
sulfotransferase family, cytosolic, 2B, member 1 |
6842 |
0.11 |
| chr11_133827302_133827588 | 0.42 |
IGSF9B |
immunoglobulin superfamily, member 9B |
565 |
0.52 |
| chr19_7456170_7456333 | 0.42 |
CTB-133G6.2 |
|
3674 |
0.16 |
| chr4_177192376_177192684 | 0.41 |
ASB5 |
ankyrin repeat and SOCS box containing 5 |
2157 |
0.35 |
| chr10_22717226_22717377 | 0.41 |
RP11-301N24.3 |
|
67200 |
0.1 |
| chrX_133147131_133147282 | 0.41 |
GPC3 |
glypican 3 |
27284 |
0.21 |
| chr7_22473101_22473341 | 0.41 |
ENSG00000263927 |
. |
53598 |
0.13 |
| chr2_28672824_28673107 | 0.41 |
PLB1 |
phospholipase B1 |
7047 |
0.22 |
| chr3_71031910_71032144 | 0.41 |
FOXP1 |
forkhead box P1 |
82050 |
0.1 |
| chr11_132910564_132910715 | 0.41 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
96976 |
0.09 |
| chr1_149911166_149911509 | 0.40 |
MTMR11 |
myotubularin related protein 11 |
2546 |
0.15 |
| chr17_2869143_2869294 | 0.40 |
CTD-3060P21.1 |
|
29 |
0.98 |
| chr10_62734565_62734716 | 0.40 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
26558 |
0.24 |
| chr3_42138838_42138989 | 0.40 |
TRAK1 |
trafficking protein, kinesin binding 1 |
6351 |
0.29 |
| chr1_63156195_63156408 | 0.40 |
RP11-230B22.1 |
|
2148 |
0.29 |
| chr17_8976590_8976741 | 0.40 |
NTN1 |
netrin 1 |
51045 |
0.15 |
| chr17_6458496_6458984 | 0.39 |
PITPNM3 |
PITPNM family member 3 |
1040 |
0.49 |
| chr18_26736738_26736889 | 0.39 |
ENSG00000212085 |
. |
39679 |
0.19 |
| chr3_55236478_55236629 | 0.39 |
LRTM1 |
leucine-rich repeats and transmembrane domains 1 |
235438 |
0.02 |
| chr11_125497290_125497441 | 0.39 |
CHEK1 |
checkpoint kinase 1 |
722 |
0.57 |
| chr18_55863472_55863729 | 0.39 |
RP11-718I15.1 |
|
672 |
0.47 |
| chr8_62621050_62621416 | 0.39 |
ASPH |
aspartate beta-hydroxylase |
5851 |
0.24 |
| chr18_47447340_47447629 | 0.39 |
MYO5B |
myosin VB |
16441 |
0.16 |
| chr9_21677527_21678485 | 0.38 |
ENSG00000244230 |
. |
21307 |
0.21 |
| chr16_29302545_29302873 | 0.38 |
RP11-231C14.6 |
|
20962 |
0.17 |
| chr21_33893367_33893849 | 0.38 |
ENSG00000252045 |
. |
17001 |
0.16 |
| chr1_76231453_76231604 | 0.38 |
RABGGTB |
Rab geranylgeranyltransferase, beta subunit |
20351 |
0.11 |
| chr16_14051400_14051742 | 0.38 |
CTD-2135D7.2 |
|
22079 |
0.22 |
| chr7_41511992_41512216 | 0.38 |
INHBA-AS1 |
INHBA antisense RNA 1 |
221410 |
0.02 |
| chr16_88298914_88299157 | 0.37 |
ZNF469 |
zinc finger protein 469 |
194844 |
0.02 |
| chr3_143550249_143550400 | 0.37 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
16976 |
0.29 |
| chr1_201818774_201819040 | 0.37 |
IPO9-AS1 |
IPO9 antisense RNA 1 |
20220 |
0.11 |
| chr18_47419130_47419281 | 0.37 |
MYO5B |
myosin VB |
10000 |
0.17 |
| chr9_140089237_140089388 | 0.37 |
TPRN |
taperin |
5245 |
0.06 |
| chrX_46838635_46838911 | 0.37 |
JADE3 |
jade family PHD finger 3 |
66542 |
0.09 |
| chr12_52835040_52835191 | 0.37 |
KRT75 |
keratin 75 |
6806 |
0.1 |
| chr6_37105912_37106091 | 0.37 |
PIM1 |
pim-1 oncogene |
31978 |
0.15 |
| chr6_150218431_150218911 | 0.37 |
RAET1E |
retinoic acid early transcript 1E |
567 |
0.64 |
| chr6_12546181_12546332 | 0.36 |
ENSG00000223321 |
. |
139243 |
0.05 |
| chr2_60743182_60743762 | 0.36 |
AC009970.1 |
|
20651 |
0.21 |
| chr8_128911768_128911919 | 0.36 |
TMEM75 |
transmembrane protein 75 |
48748 |
0.14 |
| chr9_91793130_91793737 | 0.36 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
249 |
0.95 |
| chr15_59560646_59560813 | 0.35 |
RP11-429D19.1 |
|
2632 |
0.22 |
| chr12_120242288_120242514 | 0.35 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
1214 |
0.56 |
| chr11_120211307_120211458 | 0.35 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
3436 |
0.23 |
| chr4_122612105_122612382 | 0.35 |
ANXA5 |
annexin A5 |
5874 |
0.26 |
| chr6_158034126_158034277 | 0.35 |
ZDHHC14 |
zinc finger, DHHC-type containing 14 |
20160 |
0.25 |
| chr1_205263814_205264026 | 0.35 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
26963 |
0.14 |
| chr1_115842859_115843157 | 0.34 |
RP4-663N10.1 |
|
17353 |
0.23 |
| chr9_132595482_132595633 | 0.34 |
C9orf78 |
chromosome 9 open reading frame 78 |
1997 |
0.23 |
| chr10_35927790_35928179 | 0.34 |
ENSG00000264780 |
. |
2196 |
0.26 |
| chr14_53301814_53302501 | 0.34 |
FERMT2 |
fermitin family member 2 |
29082 |
0.15 |
| chr14_65473243_65473394 | 0.34 |
RP11-840I19.5 |
|
2833 |
0.17 |
| chr1_233430570_233431212 | 0.34 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
568 |
0.83 |
| chr6_138914677_138915087 | 0.34 |
NHSL1 |
NHS-like 1 |
21205 |
0.24 |
| chr12_91503113_91503264 | 0.34 |
LUM |
lumican |
2420 |
0.34 |
| chr20_56347674_56347957 | 0.34 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
61274 |
0.13 |
| chr9_139120133_139120505 | 0.34 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
3596 |
0.22 |
| chr7_98048096_98048428 | 0.34 |
BAIAP2L1 |
BAI1-associated protein 2-like 1 |
17882 |
0.23 |
| chr10_54203606_54203757 | 0.34 |
RP11-556E13.1 |
|
117184 |
0.06 |
| chr2_224428342_224428624 | 0.33 |
SCG2 |
secretogranin II |
38519 |
0.23 |
| chr8_27471536_27471788 | 0.33 |
CLU |
clusterin |
98 |
0.97 |
| chr3_194494197_194494348 | 0.33 |
FAM43A |
family with sequence similarity 43, member A |
87650 |
0.07 |
| chr20_19973893_19974157 | 0.33 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
23735 |
0.19 |
| chr5_42641311_42641462 | 0.33 |
GHR |
growth hormone receptor |
75428 |
0.11 |
| chr14_96312809_96312960 | 0.33 |
ENSG00000263357 |
. |
5043 |
0.29 |
| chr5_17025139_17025290 | 0.33 |
BASP1 |
brain abundant, membrane attached signal protein 1 |
40493 |
0.16 |
| chr1_28431643_28431939 | 0.33 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
16584 |
0.13 |
| chr2_201630913_201631184 | 0.33 |
ENSG00000201737 |
. |
9259 |
0.15 |
| chr1_37936969_37937609 | 0.33 |
LINC01137 |
long intergenic non-protein coding RNA 1137 |
2723 |
0.2 |
| chr1_209212803_209213215 | 0.32 |
ENSG00000230937 |
. |
392469 |
0.01 |
| chr5_119741046_119741197 | 0.32 |
PRR16 |
proline rich 16 |
58852 |
0.15 |
| chr6_139870784_139870935 | 0.32 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
175102 |
0.03 |
| chr1_117542347_117542498 | 0.32 |
CD101 |
CD101 molecule |
1960 |
0.31 |
| chr15_70877228_70877484 | 0.32 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
117264 |
0.06 |
| chr12_132268438_132268589 | 0.32 |
ENSG00000200516 |
. |
17466 |
0.16 |
| chr6_116708231_116708568 | 0.32 |
DSE |
dermatan sulfate epimerase |
16289 |
0.18 |
| chr5_60524633_60524784 | 0.32 |
SMIM15 |
small integral membrane protein 15 |
66407 |
0.11 |
| chr12_69894541_69894847 | 0.32 |
FRS2 |
fibroblast growth factor receptor substrate 2 |
29909 |
0.17 |
| chr20_16800308_16800459 | 0.32 |
OTOR |
otoraplin |
71380 |
0.11 |
| chr19_30159907_30160058 | 0.32 |
PLEKHF1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
2201 |
0.36 |
| chr4_148703526_148703701 | 0.32 |
ENSG00000264274 |
. |
133 |
0.96 |
| chr4_85859566_85859717 | 0.32 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
27862 |
0.22 |
| chr5_66311217_66311678 | 0.32 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
10964 |
0.24 |
| chr8_41628467_41628645 | 0.32 |
ANK1 |
ankyrin 1, erythrocytic |
26584 |
0.16 |
| chr4_27985809_27985960 | 0.32 |
RP11-180C1.1 |
Uncharacterized protein |
418439 |
0.01 |
| chr19_18385706_18385857 | 0.32 |
KIAA1683 |
KIAA1683 |
462 |
0.67 |
| chr12_115813083_115813371 | 0.31 |
RP11-116D17.1 |
HCG2038717; Uncharacterized protein |
12410 |
0.32 |
| chr4_74308058_74308209 | 0.31 |
AFP |
alpha-fetoprotein |
6169 |
0.22 |
| chr6_112145118_112145500 | 0.31 |
FYN |
FYN oncogene related to SRC, FGR, YES |
4028 |
0.33 |
| chr6_4352330_4352530 | 0.31 |
ENSG00000201185 |
. |
75767 |
0.11 |
| chr1_147148877_147149210 | 0.31 |
ACP6 |
acid phosphatase 6, lysophosphatidic |
6425 |
0.22 |
| chr9_29039310_29039461 | 0.30 |
ENSG00000215939 |
. |
150432 |
0.05 |
| chr2_234883363_234883514 | 0.30 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
8176 |
0.21 |
| chr15_39579783_39579934 | 0.30 |
RP11-624L4.1 |
|
13774 |
0.25 |
| chr2_218784709_218784860 | 0.30 |
TNS1 |
tensin 1 |
148 |
0.97 |
| chr9_130717791_130717942 | 0.30 |
FAM102A |
family with sequence similarity 102, member A |
4865 |
0.11 |
| chr5_146783684_146783835 | 0.30 |
CTB-108O6.2 |
|
2436 |
0.31 |
| chr3_14042592_14042743 | 0.30 |
TPRXL |
tetra-peptide repeat homeobox-like |
1257 |
0.54 |
| chr17_15301686_15301923 | 0.30 |
ENSG00000239888 |
. |
9897 |
0.2 |
| chr3_64174748_64175187 | 0.30 |
PRICKLE2-AS3 |
PRICKLE2 antisense RNA 3 |
1747 |
0.38 |
| chr5_18762110_18762261 | 0.29 |
ENSG00000238674 |
. |
81821 |
0.12 |
| chr19_39853441_39853592 | 0.29 |
SAMD4B |
sterile alpha motif domain containing 4B |
6060 |
0.09 |
| chrX_34828537_34828688 | 0.29 |
FAM47B |
family with sequence similarity 47, member B |
132301 |
0.06 |
| chr12_75283505_75283691 | 0.29 |
RP11-81K13.1 |
|
131151 |
0.06 |
| chr4_187854005_187854180 | 0.29 |
ENSG00000252382 |
. |
75482 |
0.12 |
| chr11_13481327_13481478 | 0.29 |
BTBD10 |
BTB (POZ) domain containing 10 |
3326 |
0.3 |
| chr7_22898137_22898380 | 0.29 |
ENSG00000221740 |
. |
2026 |
0.38 |
| chr7_2882805_2883043 | 0.29 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
726 |
0.76 |
| chr11_7986571_7986722 | 0.29 |
NLRP10 |
NLR family, pyrin domain containing 10 |
327 |
0.83 |
| chr17_48716193_48716344 | 0.29 |
ABCC3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
4050 |
0.14 |
| chr8_48343964_48344189 | 0.29 |
SPIDR |
scaffolding protein involved in DNA repair |
8887 |
0.29 |
| chr14_88336299_88336450 | 0.28 |
GALC |
galactosylceramidase |
122808 |
0.06 |
| chr10_3270107_3270258 | 0.28 |
PITRM1 |
pitrilysin metallopeptidase 1 |
55179 |
0.15 |
| chr8_144653425_144654110 | 0.28 |
MROH6 |
maestro heat-like repeat family member 6 |
1151 |
0.21 |
| chr4_152462150_152462592 | 0.28 |
FAM160A1 |
family with sequence similarity 160, member A1 |
8207 |
0.21 |
| chr3_197044241_197044392 | 0.28 |
DLG1 |
discs, large homolog 1 (Drosophila) |
18145 |
0.15 |
| chr3_192536808_192537029 | 0.28 |
FGF12 |
fibroblast growth factor 12 |
51365 |
0.16 |
| chr18_26737213_26737364 | 0.28 |
ENSG00000212085 |
. |
39204 |
0.19 |
| chr15_93168974_93169125 | 0.28 |
FAM174B |
family with sequence similarity 174, member B |
13425 |
0.22 |
| chr6_3209808_3210009 | 0.28 |
TUBB2B |
tubulin, beta 2B class IIb |
18061 |
0.15 |
| chr12_107908945_107909258 | 0.28 |
ENSG00000222302 |
. |
28689 |
0.2 |
| chr20_20432923_20433275 | 0.28 |
RALGAPA2 |
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
60145 |
0.13 |
| chr19_58837119_58837270 | 0.28 |
ZSCAN22 |
zinc finger and SCAN domain containing 22 |
1191 |
0.24 |
| chr16_12434298_12434449 | 0.27 |
SNX29 |
sorting nexin 29 |
15737 |
0.17 |
| chr17_77022963_77023114 | 0.27 |
C1QTNF1-AS1 |
C1QTNF1 antisense RNA 1 |
645 |
0.56 |
| chr2_205660028_205660179 | 0.27 |
PARD3B |
par-3 family cell polarity regulator beta |
249380 |
0.02 |
| chr22_19564292_19564522 | 0.27 |
CLDN5 |
claudin 5 |
49339 |
0.11 |
| chr5_92414302_92414540 | 0.27 |
ENSG00000237187 |
. |
343698 |
0.01 |
| chr16_75353982_75354222 | 0.27 |
RP11-252K23.1 |
|
4844 |
0.17 |
| chr14_70059974_70060446 | 0.27 |
KIAA0247 |
KIAA0247 |
18103 |
0.2 |
| chr18_60434749_60434900 | 0.27 |
ENSG00000206746 |
. |
36259 |
0.15 |
| chr3_138153080_138153961 | 0.27 |
ESYT3 |
extended synaptotagmin-like protein 3 |
53 |
0.98 |
| chr7_33914000_33914151 | 0.27 |
BMPER |
BMP binding endothelial regulator |
30448 |
0.24 |
| chr3_170979334_170979485 | 0.27 |
TNIK |
TRAF2 and NCK interacting kinase |
35909 |
0.22 |
| chr15_85948340_85948491 | 0.27 |
ENSG00000251891 |
. |
15300 |
0.2 |
| chr4_159992618_159992885 | 0.27 |
ENSG00000206978 |
. |
7201 |
0.18 |
| chr6_159271096_159271505 | 0.27 |
C6orf99 |
chromosome 6 open reading frame 99 |
19671 |
0.16 |
| chr1_109740919_109741171 | 0.27 |
ENSG00000238310 |
. |
9425 |
0.15 |
| chr6_138571879_138572276 | 0.27 |
PBOV1 |
prostate and breast cancer overexpressed 1 |
32450 |
0.2 |
| chr15_74093075_74093391 | 0.26 |
C15orf59 |
chromosome 15 open reading frame 59 |
48145 |
0.12 |
| chr7_38885898_38886049 | 0.26 |
VPS41 |
vacuolar protein sorting 41 homolog (S. cerevisiae) |
15027 |
0.28 |
| chr4_48728692_48728843 | 0.26 |
FRYL |
FRY-like |
45579 |
0.16 |
| chr9_75771174_75771814 | 0.26 |
ANXA1 |
annexin A1 |
1076 |
0.66 |
| chr17_73680522_73680673 | 0.26 |
RP11-474I11.7 |
|
282 |
0.82 |
| chr20_25290950_25291163 | 0.26 |
ABHD12 |
abhydrolase domain containing 12 |
922 |
0.6 |
| chr3_197581656_197581918 | 0.26 |
LRCH3 |
leucine-rich repeats and calponin homology (CH) domain containing 3 |
6915 |
0.18 |
| chr17_16420060_16420464 | 0.26 |
FAM211A |
family with sequence similarity 211, member A |
24795 |
0.14 |
| chr22_40928059_40928879 | 0.26 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
1346 |
0.4 |
| chr19_45414386_45414667 | 0.26 |
APOC1 |
apolipoprotein C-I |
2978 |
0.11 |
| chr5_141700925_141701122 | 0.26 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
2724 |
0.3 |
| chr19_29521126_29521277 | 0.26 |
ENSG00000252272 |
. |
28624 |
0.24 |
| chr6_18488896_18489181 | 0.26 |
ENSG00000207775 |
. |
82977 |
0.09 |
| chr12_120637695_120638021 | 0.26 |
RPLP0 |
ribosomal protein, large, P0 |
760 |
0.43 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0021853 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.1 | 0.2 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.3 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.1 | 0.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.2 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.3 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.3 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.0 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.4 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0072111 | cell proliferation involved in kidney development(GO:0072111) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.0 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.0 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.4 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:0060294 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.5 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.7 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.0 | GO:0031579 | membrane raft organization(GO:0031579) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.0 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.2 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.4 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.0 | REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | Genes involved in Activation of NMDA receptor upon glutamate binding and postsynaptic events |