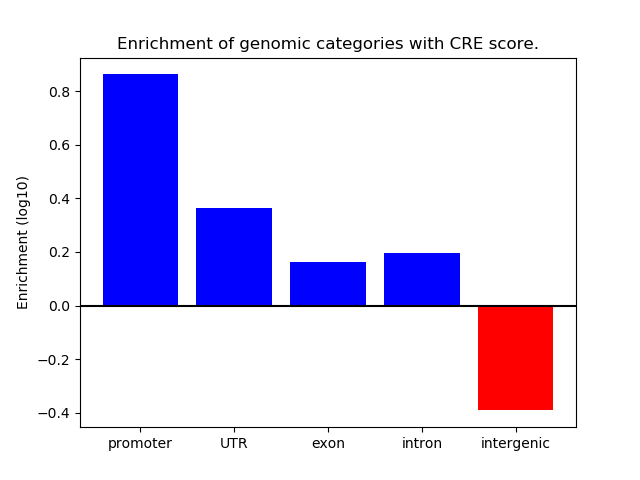
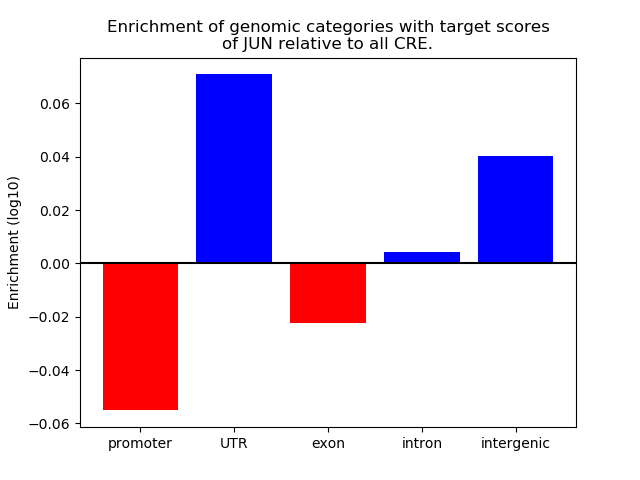
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
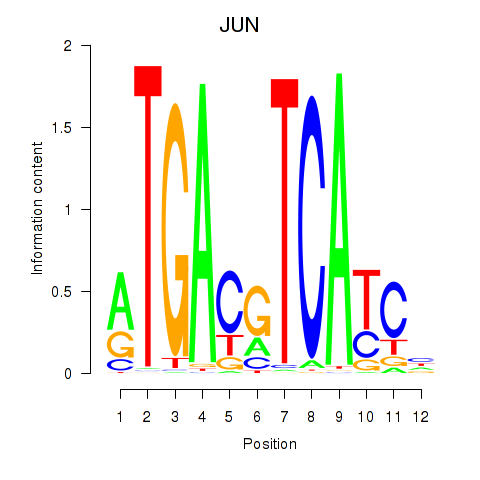
Results for JUN
Z-value: 1.05
Transcription factors associated with JUN
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JUN
|
ENSG00000177606.5 | Jun proto-oncogene, AP-1 transcription factor subunit |
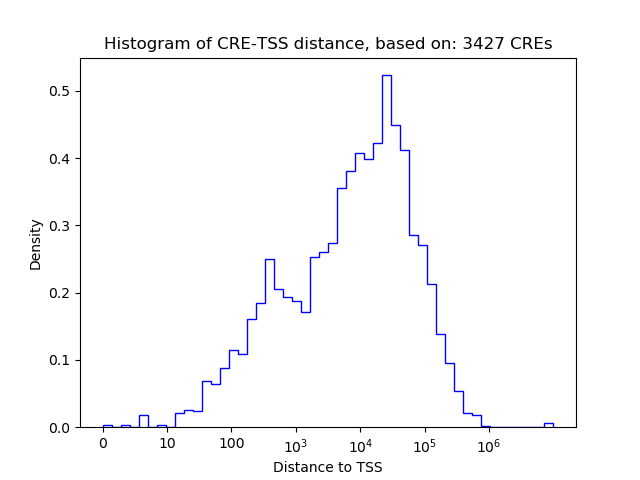
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_59485773_59486035 | JUN | 236119 | 0.019330 | -0.73 | 2.7e-02 | Click! |
| chr1_59250006_59250701 | JUN | 568 | 0.790669 | -0.72 | 3.0e-02 | Click! |
| chr1_59282219_59282390 | JUN | 32519 | 0.190534 | -0.68 | 4.3e-02 | Click! |
| chr1_59250908_59251078 | JUN | 1208 | 0.521974 | -0.65 | 5.7e-02 | Click! |
| chr1_59250755_59250906 | JUN | 1045 | 0.576408 | -0.64 | 6.2e-02 | Click! |
Activity of the JUN motif across conditions
Conditions sorted by the z-value of the JUN motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
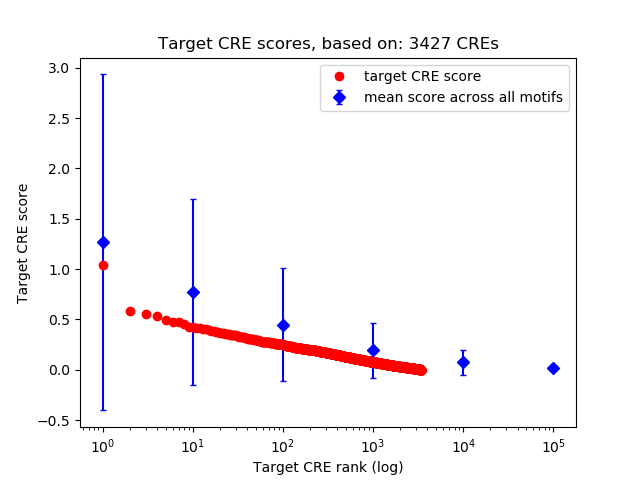
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_133022737_133022888 | 1.04 |
MUC8 |
mucin 8 |
27914 |
0.17 |
| chr17_56744273_56744616 | 0.59 |
ENSG00000199426 |
. |
386 |
0.76 |
| chr11_12695107_12695921 | 0.56 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
455 |
0.89 |
| chr8_142141679_142142152 | 0.54 |
RP11-809O17.1 |
|
1855 |
0.33 |
| chr3_128207705_128209147 | 0.50 |
RP11-475N22.4 |
|
326 |
0.79 |
| chr1_4999527_4999744 | 0.48 |
AJAP1 |
adherens junctions associated protein 1 |
284530 |
0.01 |
| chr11_77185235_77185792 | 0.47 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
167 |
0.92 |
| chr1_79471738_79472138 | 0.45 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
465 |
0.9 |
| chr6_2111373_2111600 | 0.43 |
GMDS |
GDP-mannose 4,6-dehydratase |
64739 |
0.15 |
| chr9_18976648_18977105 | 0.43 |
FAM154A |
family with sequence similarity 154, member A |
56310 |
0.1 |
| chr9_89364915_89365206 | 0.42 |
GAS1 |
growth arrest-specific 1 |
197044 |
0.03 |
| chr12_119614350_119614518 | 0.41 |
RP11-64B16.3 |
|
145 |
0.94 |
| chr9_120477806_120477957 | 0.41 |
ENSG00000201444 |
. |
7501 |
0.21 |
| chr1_219834214_219834453 | 0.40 |
ENSG00000252240 |
. |
2386 |
0.43 |
| chr19_12639322_12639473 | 0.40 |
CTD-2192J16.21 |
|
4390 |
0.14 |
| chr20_33146419_33146919 | 0.39 |
MAP1LC3A |
microtubule-associated protein 1 light chain 3 alpha |
147 |
0.95 |
| chr11_89223927_89224152 | 0.38 |
NOX4 |
NADPH oxidase 4 |
107 |
0.98 |
| chr16_57842277_57842616 | 0.38 |
CTD-2600O9.1 |
uncharacterized protein LOC388282 |
2103 |
0.23 |
| chr12_121725207_121725358 | 0.37 |
CAMKK2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
9207 |
0.21 |
| chr3_112961154_112961680 | 0.37 |
BOC |
BOC cell adhesion associated, oncogene regulated |
26219 |
0.19 |
| chr6_4491783_4491934 | 0.36 |
ENSG00000201185 |
. |
63661 |
0.14 |
| chr15_40530958_40531718 | 0.36 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
46 |
0.96 |
| chr5_131562588_131563831 | 0.36 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
138 |
0.96 |
| chr5_124194835_124194993 | 0.35 |
ZNF608 |
zinc finger protein 608 |
110414 |
0.06 |
| chr7_100464814_100466140 | 0.35 |
TRIP6 |
thyroid hormone receptor interactor 6 |
717 |
0.47 |
| chr4_169753076_169754370 | 0.35 |
RP11-635L1.3 |
|
365 |
0.61 |
| chr12_97300736_97301972 | 0.35 |
NEDD1 |
neural precursor cell expressed, developmentally down-regulated 1 |
110 |
0.98 |
| chr19_10678474_10679697 | 0.35 |
CDKN2D |
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
569 |
0.54 |
| chr3_187573527_187573678 | 0.34 |
BCL6 |
B-cell CLL/lymphoma 6 |
110087 |
0.07 |
| chr8_101522331_101522482 | 0.34 |
KB-1615E4.3 |
|
17650 |
0.16 |
| chr8_2199973_2200269 | 0.34 |
MYOM2 |
myomesin 2 |
206937 |
0.03 |
| chr1_209846224_209846375 | 0.34 |
RP1-28O10.1 |
|
2293 |
0.2 |
| chr7_37956036_37956797 | 0.33 |
SFRP4 |
secreted frizzled-related protein 4 |
101 |
0.97 |
| chr6_47276454_47277750 | 0.33 |
TNFRSF21 |
tumor necrosis factor receptor superfamily, member 21 |
539 |
0.86 |
| chrX_10126292_10126538 | 0.33 |
CLCN4 |
chloride channel, voltage-sensitive 4 |
73 |
0.98 |
| chr9_127024177_127024426 | 0.33 |
NEK6 |
NIMA-related kinase 6 |
549 |
0.47 |
| chr17_38497502_38498255 | 0.32 |
RARA |
retinoic acid receptor, alpha |
238 |
0.84 |
| chr1_167624325_167624476 | 0.32 |
RP3-455J7.4 |
|
24489 |
0.17 |
| chr21_45139141_45140358 | 0.32 |
PDXK |
pyridoxal (pyridoxine, vitamin B6) kinase |
756 |
0.66 |
| chr2_9621262_9621413 | 0.31 |
IAH1 |
isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae) |
6153 |
0.18 |
| chr6_30523511_30524213 | 0.31 |
GNL1 |
guanine nucleotide binding protein-like 1 |
3 |
0.92 |
| chr2_180214031_180214182 | 0.31 |
SESTD1 |
SEC14 and spectrin domains 1 |
84589 |
0.1 |
| chr18_74779349_74779519 | 0.31 |
MBP |
myelin basic protein |
37783 |
0.2 |
| chr1_240161229_240161618 | 0.31 |
FMN2 |
formin 2 |
16225 |
0.27 |
| chr5_126147492_126147682 | 0.30 |
LMNB1 |
lamin B1 |
34706 |
0.19 |
| chr3_151102768_151102919 | 0.30 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
243 |
0.93 |
| chr2_28939308_28939670 | 0.30 |
AC097724.3 |
|
19486 |
0.15 |
| chr1_155734748_155734899 | 0.30 |
DAP3 |
death associated protein 3 |
28020 |
0.13 |
| chr16_66638822_66640143 | 0.30 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
58 |
0.95 |
| chr10_14920855_14921866 | 0.30 |
SUV39H2 |
suppressor of variegation 3-9 homolog 2 (Drosophila) |
439 |
0.82 |
| chr1_101334241_101334590 | 0.30 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
25805 |
0.15 |
| chr9_35489468_35490886 | 0.30 |
RUSC2 |
RUN and SH3 domain containing 2 |
53 |
0.97 |
| chr1_46379145_46379296 | 0.29 |
MAST2 |
microtubule associated serine/threonine kinase 2 |
40 |
0.98 |
| chr2_192764216_192764367 | 0.29 |
AC098617.1 |
|
22981 |
0.23 |
| chr1_203631994_203632193 | 0.29 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
19844 |
0.21 |
| chr15_48469342_48470564 | 0.28 |
MYEF2 |
myelin expression factor 2 |
597 |
0.66 |
| chr7_121951944_121952095 | 0.28 |
FEZF1 |
FEZ family zinc finger 1 |
1274 |
0.46 |
| chr2_204414201_204414352 | 0.28 |
RAPH1 |
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
14143 |
0.28 |
| chr17_48367766_48367999 | 0.28 |
RP11-893F2.9 |
|
2666 |
0.19 |
| chr3_185903259_185903649 | 0.28 |
ETV5 |
ets variant 5 |
75347 |
0.1 |
| chr3_184056155_184056581 | 0.28 |
FAM131A |
family with sequence similarity 131, member A |
249 |
0.84 |
| chr22_38656697_38656971 | 0.28 |
TMEM184B |
transmembrane protein 184B |
11836 |
0.11 |
| chr2_40264407_40264558 | 0.28 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
5008 |
0.35 |
| chr1_186798918_186799215 | 0.28 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
944 |
0.73 |
| chr18_73522464_73522615 | 0.28 |
SMIM21 |
small integral membrane protein 21 |
382881 |
0.01 |
| chr5_32033164_32033315 | 0.28 |
CTD-2152M20.2 |
|
88808 |
0.07 |
| chr13_32610061_32610212 | 0.28 |
FRY-AS1 |
FRY antisense RNA 1 |
4360 |
0.25 |
| chr5_136599797_136599948 | 0.28 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
49355 |
0.19 |
| chr18_3262315_3263346 | 0.27 |
MYL12B |
myosin, light chain 12B, regulatory |
124 |
0.92 |
| chrX_154027559_154027756 | 0.27 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
6015 |
0.13 |
| chr3_89163237_89163857 | 0.27 |
EPHA3 |
EPH receptor A3 |
6721 |
0.35 |
| chr7_993167_993948 | 0.27 |
ADAP1 |
ArfGAP with dual PH domains 1 |
776 |
0.55 |
| chrX_14130141_14130292 | 0.27 |
ENSG00000264331 |
. |
36206 |
0.21 |
| chr22_45664060_45664211 | 0.27 |
UPK3A |
uroplakin 3A |
16728 |
0.16 |
| chr7_55573864_55574015 | 0.27 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
9831 |
0.26 |
| chr1_2187001_2187152 | 0.27 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
26942 |
0.11 |
| chr8_25056770_25056921 | 0.27 |
DOCK5 |
dedicator of cytokinesis 5 |
14465 |
0.23 |
| chr3_11680615_11680766 | 0.27 |
VGLL4 |
vestigial like 4 (Drosophila) |
4708 |
0.24 |
| chr7_33088447_33088598 | 0.26 |
ENSG00000241420 |
. |
2820 |
0.22 |
| chr1_161515451_161515602 | 0.26 |
FCGR3A |
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
4053 |
0.15 |
| chr3_120168709_120169860 | 0.26 |
FSTL1 |
follistatin-like 1 |
554 |
0.84 |
| chr4_173747868_173748019 | 0.26 |
ENSG00000241652 |
. |
314851 |
0.01 |
| chr10_3818147_3818693 | 0.26 |
RP11-184A2.2 |
|
8313 |
0.21 |
| chr8_38722630_38723350 | 0.26 |
RP11-723D22.3 |
|
19033 |
0.16 |
| chr4_186346547_186347774 | 0.26 |
UFSP2 |
UFM1-specific peptidase 2 |
21 |
0.76 |
| chr18_73350794_73351073 | 0.26 |
SMIM21 |
small integral membrane protein 21 |
211275 |
0.02 |
| chr18_29629892_29630043 | 0.26 |
ENSG00000265063 |
. |
10985 |
0.14 |
| chr18_74333391_74333920 | 0.26 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
92781 |
0.08 |
| chr6_132520617_132520826 | 0.26 |
ENSG00000265669 |
. |
84318 |
0.1 |
| chr4_2010754_2011490 | 0.26 |
NELFA |
negative elongation factor complex member A |
398 |
0.81 |
| chrX_42106662_42106813 | 0.26 |
ENSG00000206896 |
. |
73099 |
0.12 |
| chr1_9256722_9257085 | 0.26 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
37931 |
0.13 |
| chr8_57471403_57471760 | 0.26 |
RP11-17A4.2 |
|
69924 |
0.12 |
| chr11_61722707_61723219 | 0.26 |
BEST1 |
bestrophin 1 |
334 |
0.82 |
| chr2_101945070_101945221 | 0.26 |
ENSG00000264857 |
. |
19233 |
0.16 |
| chr8_129874103_129874314 | 0.25 |
ENSG00000221351 |
. |
42168 |
0.22 |
| chr13_48718622_48718773 | 0.25 |
MED4 |
mediator complex subunit 4 |
49430 |
0.14 |
| chr7_41740779_41741576 | 0.25 |
INHBA |
inhibin, beta A |
970 |
0.56 |
| chr20_43988486_43988637 | 0.25 |
SYS1 |
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
2016 |
0.21 |
| chr10_118764443_118765166 | 0.25 |
KIAA1598 |
KIAA1598 |
72 |
0.98 |
| chr2_74097379_74097530 | 0.25 |
ACTG2 |
actin, gamma 2, smooth muscle, enteric |
22640 |
0.15 |
| chr17_79485709_79486744 | 0.25 |
RP13-766D20.2 |
|
160 |
0.9 |
| chr5_154026662_154027279 | 0.25 |
ENSG00000221552 |
. |
38366 |
0.13 |
| chr7_4722786_4723588 | 0.24 |
FOXK1 |
forkhead box K1 |
1247 |
0.49 |
| chr6_11795100_11795449 | 0.24 |
ADTRP |
androgen-dependent TFPI-regulating protein |
12005 |
0.28 |
| chr2_172944774_172945201 | 0.24 |
DLX1 |
distal-less homeobox 1 |
4481 |
0.24 |
| chr18_61559668_61559819 | 0.24 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
1952 |
0.31 |
| chr17_37052701_37052852 | 0.24 |
RP1-56K13.2 |
|
8077 |
0.11 |
| chr2_46767914_46768065 | 0.24 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
1707 |
0.26 |
| chr14_101294998_101295174 | 0.24 |
AL117190.2 |
|
451 |
0.51 |
| chr13_99227754_99228977 | 0.24 |
STK24 |
serine/threonine kinase 24 |
752 |
0.54 |
| chr3_99964720_99964871 | 0.24 |
TBC1D23 |
TBC1 domain family, member 23 |
15049 |
0.17 |
| chr8_126557270_126557537 | 0.24 |
ENSG00000266452 |
. |
100596 |
0.08 |
| chr12_10902635_10902853 | 0.24 |
YBX3 |
Y box binding protein 3 |
26833 |
0.13 |
| chr1_242097620_242097936 | 0.24 |
EXO1 |
exonuclease 1 |
55436 |
0.13 |
| chr2_7144548_7144834 | 0.24 |
RNF144A |
ring finger protein 144A |
7620 |
0.26 |
| chr3_182977447_182977598 | 0.23 |
ENSG00000202502 |
. |
1318 |
0.46 |
| chr4_6887603_6887754 | 0.23 |
TBC1D14 |
TBC1 domain family, member 14 |
23291 |
0.18 |
| chr10_97505576_97505727 | 0.23 |
ENTPD1 |
ectonucleoside triphosphate diphosphohydrolase 1 |
9758 |
0.23 |
| chr10_23983047_23983671 | 0.23 |
KIAA1217 |
KIAA1217 |
316 |
0.94 |
| chr17_53342049_53343259 | 0.23 |
HLF |
hepatic leukemia factor |
281 |
0.93 |
| chr12_4699544_4699695 | 0.23 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
57 |
0.97 |
| chr4_40669941_40670092 | 0.23 |
RBM47 |
RNA binding motif protein 47 |
37124 |
0.18 |
| chr2_1609000_1609151 | 0.23 |
AC144450.1 |
|
14810 |
0.26 |
| chr13_51253017_51253168 | 0.23 |
DLEU7-AS1 |
DLEU7 antisense RNA 1 |
128900 |
0.05 |
| chr2_64864648_64864996 | 0.23 |
SERTAD2 |
SERTA domain containing 2 |
16225 |
0.22 |
| chr5_120114458_120114855 | 0.23 |
ENSG00000222609 |
. |
68121 |
0.14 |
| chr17_14507939_14508129 | 0.23 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
303634 |
0.01 |
| chr3_42055388_42056674 | 0.23 |
ULK4 |
unc-51 like kinase 4 |
52109 |
0.14 |
| chr12_6178520_6178671 | 0.23 |
ENSG00000240533 |
. |
26541 |
0.19 |
| chrX_24043946_24044693 | 0.23 |
KLHL15 |
kelch-like family member 15 |
984 |
0.59 |
| chr19_2049519_2050886 | 0.23 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
650 |
0.55 |
| chr7_130712091_130712242 | 0.22 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
25143 |
0.19 |
| chr7_99755308_99756343 | 0.22 |
C7orf43 |
chromosome 7 open reading frame 43 |
182 |
0.82 |
| chr12_25705930_25706081 | 0.22 |
IFLTD1 |
intermediate filament tail domain containing 1 |
173 |
0.97 |
| chr5_131111835_131111986 | 0.22 |
FNIP1 |
folliculin interacting protein 1 |
20704 |
0.2 |
| chr4_99443689_99443840 | 0.22 |
RP11-724M22.1 |
|
26249 |
0.24 |
| chr19_45242197_45242348 | 0.22 |
BCL3 |
B-cell CLL/lymphoma 3 |
9532 |
0.11 |
| chr13_58208642_58209078 | 0.22 |
PCDH17 |
protocadherin 17 |
2205 |
0.48 |
| chr6_100316349_100316522 | 0.22 |
MCHR2-AS1 |
MCHR2 antisense RNA 1 |
125385 |
0.05 |
| chr17_5371546_5372510 | 0.22 |
DHX33 |
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
40 |
0.96 |
| chr10_62175082_62175240 | 0.22 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
25673 |
0.27 |
| chr8_132054499_132054781 | 0.22 |
ADCY8 |
adenylate cyclase 8 (brain) |
32 |
0.99 |
| chr2_166325831_166326039 | 0.22 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
222 |
0.96 |
| chr18_10131788_10132277 | 0.22 |
ENSG00000263630 |
. |
126838 |
0.05 |
| chr13_48073693_48073892 | 0.22 |
ENSG00000244521 |
. |
40423 |
0.23 |
| chr11_93275392_93276314 | 0.22 |
SMCO4 |
single-pass membrane protein with coiled-coil domains 4 |
668 |
0.7 |
| chr16_75287131_75287498 | 0.22 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
1536 |
0.31 |
| chr10_64392557_64392708 | 0.22 |
ZNF365 |
zinc finger protein 365 |
10901 |
0.24 |
| chrX_133688697_133688848 | 0.22 |
ENSG00000223749 |
. |
8031 |
0.12 |
| chr17_29814991_29815971 | 0.22 |
RAB11FIP4 |
RAB11 family interacting protein 4 (class II) |
355 |
0.83 |
| chr18_77866868_77867381 | 0.22 |
ADNP2 |
ADNP homeobox 2 |
53 |
0.98 |
| chr11_31834234_31835305 | 0.21 |
PAX6 |
paired box 6 |
295 |
0.8 |
| chr6_90695271_90695484 | 0.21 |
ENSG00000222078 |
. |
15848 |
0.19 |
| chr16_11667077_11667228 | 0.21 |
LITAF |
lipopolysaccharide-induced TNF factor |
13077 |
0.18 |
| chr3_185541036_185541485 | 0.21 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
1540 |
0.46 |
| chr2_145265495_145265831 | 0.21 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
9452 |
0.25 |
| chr4_30823764_30823915 | 0.21 |
RP11-619J20.1 |
|
28247 |
0.26 |
| chr1_221204797_221204948 | 0.21 |
HLX |
H2.0-like homeobox |
150288 |
0.04 |
| chr2_179395534_179395685 | 0.21 |
TTN-AS1 |
TTN antisense RNA 1 |
67 |
0.96 |
| chr10_45956574_45956725 | 0.21 |
RP11-67C2.2 |
|
8080 |
0.26 |
| chr19_1019836_1021109 | 0.21 |
TMEM259 |
transmembrane protein 259 |
373 |
0.62 |
| chr4_119684707_119684858 | 0.21 |
SEC24D |
SEC24 family member D |
4985 |
0.3 |
| chr9_95724300_95724451 | 0.21 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
1868 |
0.38 |
| chr2_103104653_103104941 | 0.21 |
SLC9A4 |
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
15035 |
0.18 |
| chr1_184836110_184836261 | 0.21 |
ENSG00000252222 |
. |
45562 |
0.15 |
| chr7_30174028_30174462 | 0.21 |
MTURN |
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
181 |
0.94 |
| chr19_33686404_33686657 | 0.21 |
LRP3 |
low density lipoprotein receptor-related protein 3 |
1040 |
0.41 |
| chr17_79884123_79885283 | 0.21 |
MAFG |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
881 |
0.22 |
| chr11_11418107_11418277 | 0.21 |
CSNK2A3 |
casein kinase 2, alpha 3 polypeptide |
43288 |
0.17 |
| chr12_106229481_106229632 | 0.21 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
248255 |
0.02 |
| chr4_122060815_122060966 | 0.21 |
TNIP3 |
TNFAIP3 interacting protein 3 |
2164 |
0.37 |
| chr14_89412870_89413171 | 0.21 |
TTC8 |
tetratricopeptide repeat domain 8 |
105148 |
0.07 |
| chr19_40871912_40872253 | 0.21 |
PLD3 |
phospholipase D family, member 3 |
181 |
0.9 |
| chr1_219635246_219635397 | 0.21 |
ENSG00000252240 |
. |
201398 |
0.03 |
| chr7_151433344_151433930 | 0.21 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
237 |
0.95 |
| chr17_72453956_72454107 | 0.21 |
CD300A |
CD300a molecule |
8524 |
0.14 |
| chr6_42283591_42283839 | 0.21 |
ENSG00000221252 |
. |
96419 |
0.06 |
| chr8_28570812_28570963 | 0.21 |
EXTL3 |
exostosin-like glycosyltransferase 3 |
3012 |
0.26 |
| chr11_12237907_12238058 | 0.21 |
RP11-265D17.2 |
|
46738 |
0.13 |
| chr5_176328832_176328983 | 0.20 |
HK3 |
hexokinase 3 (white cell) |
2574 |
0.31 |
| chr19_58723136_58723287 | 0.20 |
ZNF544 |
zinc finger protein 544 |
16749 |
0.11 |
| chr7_8054646_8054797 | 0.20 |
AC006042.7 |
|
45187 |
0.13 |
| chr1_112147952_112148103 | 0.20 |
RAP1A |
RAP1A, member of RAS oncogene family |
14372 |
0.17 |
| chr12_106704608_106704759 | 0.20 |
CKAP4 |
cytoskeleton-associated protein 4 |
6626 |
0.18 |
| chr3_185911353_185912009 | 0.20 |
ETV5 |
ets variant 5 |
83574 |
0.09 |
| chrX_71321784_71322102 | 0.20 |
RGAG4 |
retrotransposon gag domain containing 4 |
29735 |
0.14 |
| chr8_59057981_59058941 | 0.20 |
FAM110B |
family with sequence similarity 110, member B |
151348 |
0.04 |
| chr1_247611615_247612128 | 0.20 |
OR2B11 |
olfactory receptor, family 2, subfamily B, member 11 |
3437 |
0.2 |
| chr18_76828422_76829072 | 0.20 |
ATP9B |
ATPase, class II, type 9B |
538 |
0.84 |
| chr11_308095_309459 | 0.20 |
IFITM2 |
interferon induced transmembrane protein 2 |
369 |
0.66 |
| chr17_53573866_53574017 | 0.20 |
ENSG00000200107 |
. |
55230 |
0.15 |
| chr3_119069123_119069274 | 0.20 |
ENSG00000207310 |
. |
8411 |
0.18 |
| chr15_64643443_64643594 | 0.20 |
CSNK1G1 |
casein kinase 1, gamma 1 |
4854 |
0.14 |
| chr4_153561915_153562066 | 0.20 |
RP11-768B22.2 |
|
25530 |
0.19 |
| chr2_71300827_71300978 | 0.20 |
NAGK |
N-acetylglucosamine kinase |
235 |
0.87 |
| chr11_109656621_109656772 | 0.20 |
RP11-708B6.2 |
|
202063 |
0.03 |
| chr12_81042956_81043107 | 0.20 |
PTPRQ |
protein tyrosine phosphatase, receptor type, Q |
17030 |
0.21 |
| chr5_52218355_52218506 | 0.20 |
ITGA2 |
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
66726 |
0.11 |
| chr14_37050633_37050993 | 0.20 |
NKX2-8 |
NK2 homeobox 8 |
999 |
0.51 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.3 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.2 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.2 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.1 | 0.2 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.2 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0006991 | response to sterol depletion(GO:0006991) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0045064 | T-helper 2 cell differentiation(GO:0045064) |
| 0.0 | 0.2 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.5 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.4 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.3 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.4 | GO:0006984 | ER-nucleus signaling pathway(GO:0006984) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.2 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.0 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:1904376 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.0 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.0 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.0 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.0 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.0 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0003079 | obsolete positive regulation of natriuresis(GO:0003079) |
| 0.0 | 0.2 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.2 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.0 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.0 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |