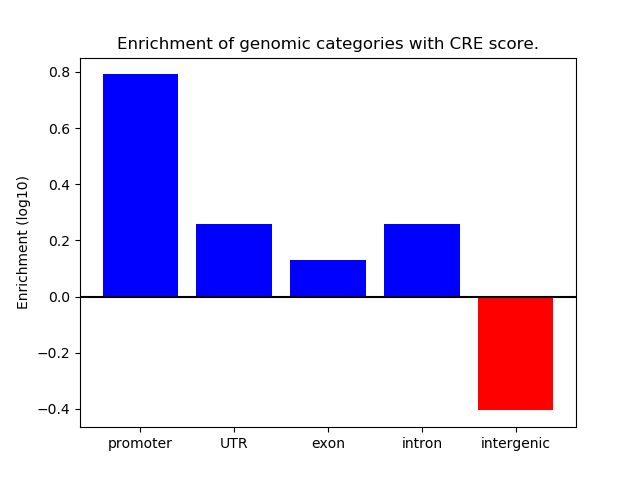
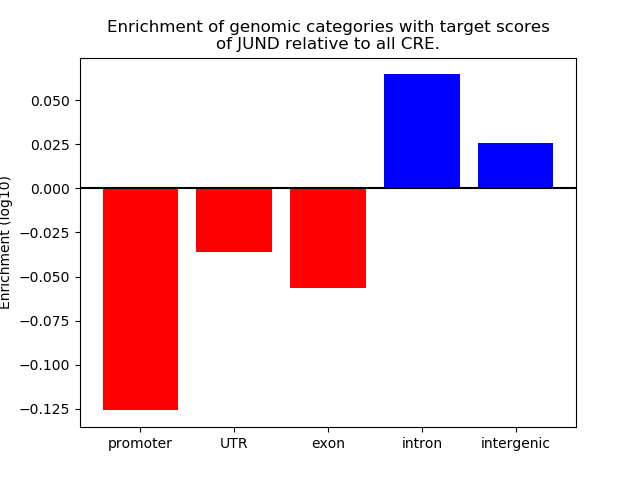
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
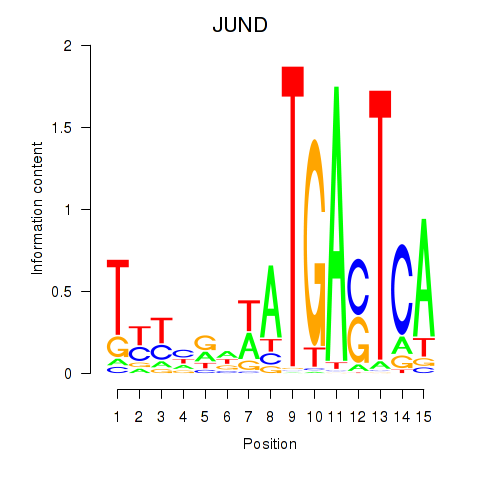
Results for JUND
Z-value: 1.73
Transcription factors associated with JUND
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JUND
|
ENSG00000130522.4 | JunD proto-oncogene, AP-1 transcription factor subunit |
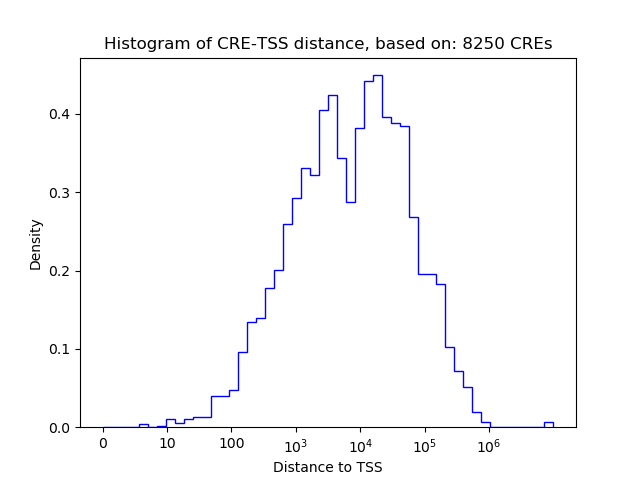
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_18388821_18388972 | JUND | 2843 | 0.131727 | 0.48 | 1.9e-01 | Click! |
| chr19_18389064_18389254 | JUND | 2580 | 0.140432 | 0.39 | 3.0e-01 | Click! |
| chr19_18389866_18390017 | JUND | 1798 | 0.187139 | 0.19 | 6.2e-01 | Click! |
| chr19_18391872_18392200 | JUND | 297 | 0.725285 | -0.18 | 6.4e-01 | Click! |
| chr19_18390478_18391041 | JUND | 980 | 0.341134 | 0.13 | 7.4e-01 | Click! |
Activity of the JUND motif across conditions
Conditions sorted by the z-value of the JUND motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
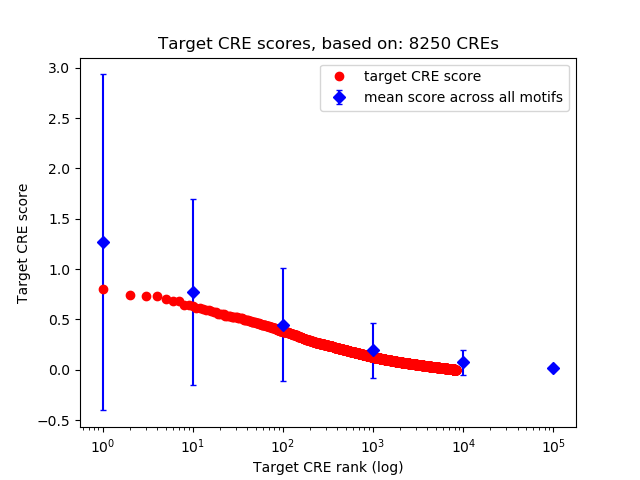
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_33373915_33374211 | 0.80 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
4693 |
0.13 |
| chr5_35857058_35857643 | 0.74 |
IL7R |
interleukin 7 receptor |
356 |
0.89 |
| chr11_118794237_118794443 | 0.73 |
BCL9L |
B-cell CLL/lymphoma 9-like |
4727 |
0.09 |
| chr14_22971834_22972057 | 0.73 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
15774 |
0.09 |
| chr4_122164507_122165136 | 0.71 |
TNIP3 |
TNFAIP3 interacting protein 3 |
16200 |
0.22 |
| chr18_9117310_9117583 | 0.69 |
RP11-143J12.3 |
|
1571 |
0.34 |
| chr4_143325352_143326244 | 0.68 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
26614 |
0.28 |
| chr6_139852295_139852446 | 0.65 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
156613 |
0.04 |
| chr5_156616289_156616440 | 0.64 |
ITK |
IL2-inducible T-cell kinase |
8527 |
0.13 |
| chr5_151907362_151907513 | 0.64 |
NMUR2 |
neuromedin U receptor 2 |
122597 |
0.07 |
| chr1_160489868_160490211 | 0.62 |
SLAMF6 |
SLAM family member 6 |
2993 |
0.18 |
| chr1_160767367_160767518 | 0.61 |
LY9 |
lymphocyte antigen 9 |
1465 |
0.39 |
| chr2_231789411_231789754 | 0.60 |
GPR55 |
G protein-coupled receptor 55 |
359 |
0.85 |
| chr12_92172716_92172989 | 0.59 |
C12orf79 |
chromosome 12 open reading frame 79 |
357945 |
0.01 |
| chr22_31335429_31335580 | 0.59 |
MORC2 |
MORC family CW-type zinc finger 2 |
6623 |
0.16 |
| chr11_128376572_128377141 | 0.58 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
1567 |
0.47 |
| chr1_117297247_117297398 | 0.58 |
CD2 |
CD2 molecule |
233 |
0.94 |
| chr9_73027094_73027481 | 0.57 |
KLF9 |
Kruppel-like factor 9 |
2253 |
0.41 |
| chr12_21606326_21606605 | 0.56 |
PYROXD1 |
pyridine nucleotide-disulphide oxidoreductase domain 1 |
15616 |
0.17 |
| chr4_151540397_151540583 | 0.56 |
ENSG00000221235 |
. |
20528 |
0.2 |
| chr6_90993958_90994705 | 0.55 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
12130 |
0.24 |
| chr12_51418457_51418700 | 0.55 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
48 |
0.96 |
| chr1_12509705_12509856 | 0.54 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
15215 |
0.22 |
| chr1_154299291_154299442 | 0.54 |
ATP8B2 |
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
910 |
0.38 |
| chr22_37545294_37545803 | 0.53 |
IL2RB |
interleukin 2 receptor, beta |
482 |
0.7 |
| chr18_7958694_7958932 | 0.53 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
11861 |
0.23 |
| chr4_143477803_143478104 | 0.53 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
3869 |
0.39 |
| chr9_130732789_130732970 | 0.53 |
FAM102A |
family with sequence similarity 102, member A |
9913 |
0.11 |
| chr3_179372008_179372371 | 0.53 |
USP13 |
ubiquitin specific peptidase 13 (isopeptidase T-3) |
708 |
0.65 |
| chr2_223654006_223654270 | 0.53 |
ACSL3 |
acyl-CoA synthetase long-chain family member 3 |
71514 |
0.1 |
| chr12_45625038_45625189 | 0.52 |
ANO6 |
anoctamin 6 |
15210 |
0.28 |
| chr4_26024061_26024293 | 0.51 |
SMIM20 |
small integral membrane protein 20 |
108246 |
0.07 |
| chr5_52336982_52337335 | 0.51 |
CTD-2366F13.2 |
|
48140 |
0.12 |
| chr12_96592768_96592919 | 0.51 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
4451 |
0.24 |
| chr20_3913213_3913723 | 0.51 |
ENSG00000199024 |
. |
15327 |
0.15 |
| chr18_3459636_3459787 | 0.50 |
TGIF1 |
TGFB-induced factor homeobox 1 |
4299 |
0.25 |
| chr20_48510567_48510837 | 0.50 |
SPATA2 |
spermatogenesis associated 2 |
19574 |
0.13 |
| chr18_9118459_9118745 | 0.50 |
RP11-143J12.3 |
|
2727 |
0.22 |
| chr21_15906632_15906783 | 0.50 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
11955 |
0.25 |
| chr3_187520878_187521148 | 0.49 |
BCL6 |
B-cell CLL/lymphoma 6 |
57498 |
0.14 |
| chr6_42391109_42391260 | 0.49 |
ENSG00000221252 |
. |
11050 |
0.21 |
| chr10_6183056_6183884 | 0.48 |
PFKFB3 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
3411 |
0.18 |
| chr13_64046885_64047036 | 0.48 |
LINC00395 |
long intergenic non-protein coding RNA 395 |
244191 |
0.02 |
| chr8_17784028_17784466 | 0.48 |
PCM1 |
pericentriolar material 1 |
1994 |
0.33 |
| chr13_50940025_50940517 | 0.48 |
ENSG00000221198 |
. |
2898 |
0.4 |
| chr15_57214479_57214630 | 0.48 |
TCF12 |
transcription factor 12 |
3208 |
0.26 |
| chr2_12861826_12862019 | 0.47 |
TRIB2 |
tribbles pseudokinase 2 |
3558 |
0.31 |
| chr20_62368732_62368892 | 0.47 |
LIME1 |
Lck interacting transmembrane adaptor 1 |
175 |
0.82 |
| chr5_125759997_125760148 | 0.47 |
GRAMD3 |
GRAM domain containing 3 |
922 |
0.67 |
| chr2_161347323_161347474 | 0.47 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
2631 |
0.39 |
| chr9_80454914_80455065 | 0.47 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
17074 |
0.29 |
| chr3_97591431_97591802 | 0.46 |
CRYBG3 |
beta-gamma crystallin domain containing 3 |
4203 |
0.29 |
| chr14_99728002_99728348 | 0.46 |
AL109767.1 |
|
1110 |
0.55 |
| chr15_57199778_57200042 | 0.46 |
ZNF280D |
zinc finger protein 280D |
10859 |
0.22 |
| chr3_69137280_69137433 | 0.45 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
3184 |
0.19 |
| chr12_27975145_27975352 | 0.45 |
ENSG00000201612 |
. |
15925 |
0.14 |
| chr1_239711433_239711584 | 0.45 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
80865 |
0.11 |
| chr10_121464981_121465132 | 0.45 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
20553 |
0.2 |
| chr12_50614114_50614771 | 0.45 |
RP3-405J10.4 |
|
1055 |
0.35 |
| chr6_106428787_106428953 | 0.45 |
ENSG00000200198 |
. |
76622 |
0.11 |
| chr17_68133569_68133739 | 0.45 |
AC002539.1 |
|
8654 |
0.19 |
| chr2_205660028_205660179 | 0.44 |
PARD3B |
par-3 family cell polarity regulator beta |
249380 |
0.02 |
| chr6_106550011_106550199 | 0.44 |
RP1-134E15.3 |
|
2090 |
0.34 |
| chr3_173342884_173343035 | 0.44 |
NLGN1 |
neuroligin 1 |
20570 |
0.29 |
| chr10_12762743_12762973 | 0.44 |
ENSG00000221331 |
. |
4494 |
0.33 |
| chrY_18612478_18612629 | 0.44 |
ENSG00000252323 |
. |
164284 |
0.04 |
| chr14_99706254_99706405 | 0.43 |
AL109767.1 |
|
22956 |
0.19 |
| chr10_531510_531790 | 0.43 |
RP11-490E15.2 |
|
48149 |
0.14 |
| chr15_26095881_26096079 | 0.43 |
ENSG00000266517 |
. |
2008 |
0.32 |
| chr1_226919354_226919568 | 0.43 |
ITPKB |
inositol-trisphosphate 3-kinase B |
5698 |
0.25 |
| chr7_80589868_80590019 | 0.43 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
38268 |
0.23 |
| chr11_58341076_58341416 | 0.42 |
LPXN |
leupaxin |
2088 |
0.26 |
| chr18_60253252_60253584 | 0.42 |
ZCCHC2 |
zinc finger, CCHC domain containing 2 |
46671 |
0.15 |
| chr16_50779002_50779153 | 0.42 |
RP11-327F22.1 |
|
2351 |
0.16 |
| chr9_5510404_5511024 | 0.42 |
PDCD1LG2 |
programmed cell death 1 ligand 2 |
144 |
0.97 |
| chr11_62308687_62309106 | 0.42 |
RP11-864I4.3 |
|
4112 |
0.1 |
| chr12_47606378_47606570 | 0.42 |
PCED1B |
PC-esterase domain containing 1B |
3578 |
0.27 |
| chr2_85165728_85166011 | 0.42 |
KCMF1 |
potassium channel modulatory factor 1 |
32347 |
0.14 |
| chr13_76867689_76867887 | 0.42 |
ENSG00000243274 |
. |
158721 |
0.04 |
| chr17_8294973_8295124 | 0.42 |
RNF222 |
ring finger protein 222 |
1759 |
0.2 |
| chr7_43709455_43709606 | 0.41 |
COA1 |
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
19635 |
0.2 |
| chr3_12436703_12437398 | 0.41 |
PPARG |
peroxisome proliferator-activated receptor gamma |
44079 |
0.15 |
| chr1_178096168_178096398 | 0.41 |
RASAL2 |
RAS protein activator like 2 |
33007 |
0.24 |
| chr10_3239818_3240224 | 0.41 |
PITRM1 |
pitrilysin metallopeptidase 1 |
25018 |
0.22 |
| chr17_18089190_18089923 | 0.41 |
RP11-258F1.1 |
|
1489 |
0.25 |
| chr1_205263608_205263759 | 0.41 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
27200 |
0.13 |
| chr2_106363775_106364396 | 0.40 |
NCK2 |
NCK adaptor protein 2 |
1897 |
0.48 |
| chr17_78735104_78735701 | 0.39 |
RP11-28G8.1 |
|
44030 |
0.16 |
| chr3_128898182_128898356 | 0.39 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
4473 |
0.15 |
| chr2_102720712_102721707 | 0.39 |
IL1R1 |
interleukin 1 receptor, type I |
180 |
0.96 |
| chrY_16456835_16457019 | 0.39 |
ENSG00000266220 |
. |
92756 |
0.09 |
| chr14_68717009_68717160 | 0.39 |
ENSG00000243546 |
. |
13828 |
0.27 |
| chr20_30948041_30948438 | 0.39 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
689 |
0.71 |
| chr17_75641513_75641796 | 0.39 |
SEPT9 |
septin 9 |
163659 |
0.03 |
| chr14_23015461_23015710 | 0.39 |
AE000662.92 |
Uncharacterized protein |
9949 |
0.11 |
| chr14_69130333_69130485 | 0.39 |
CTD-2325P2.3 |
|
21873 |
0.2 |
| chr3_67040557_67040708 | 0.38 |
KBTBD8 |
kelch repeat and BTB (POZ) domain containing 8 |
8095 |
0.34 |
| chr3_50354430_50354581 | 0.38 |
HYAL2 |
hyaluronoglucosaminidase 2 |
3874 |
0.08 |
| chr6_112145118_112145500 | 0.38 |
FYN |
FYN oncogene related to SRC, FGR, YES |
4028 |
0.33 |
| chr1_116919885_116920582 | 0.38 |
ATP1A1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
3744 |
0.2 |
| chr3_21788888_21789039 | 0.38 |
ZNF385D |
zinc finger protein 385D |
3964 |
0.35 |
| chr6_136968777_136969043 | 0.38 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
8689 |
0.18 |
| chr6_119398292_119398443 | 0.38 |
FAM184A |
family with sequence similarity 184, member A |
1170 |
0.49 |
| chr1_111215065_111215351 | 0.38 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
2447 |
0.28 |
| chr6_139467434_139467700 | 0.38 |
HECA |
headcase homolog (Drosophila) |
11318 |
0.25 |
| chr8_102914659_102914810 | 0.38 |
ENSG00000238372 |
. |
55146 |
0.15 |
| chrX_78196699_78196850 | 0.38 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
4055 |
0.37 |
| chr1_231762136_231762321 | 0.37 |
DISC1 |
disrupted in schizophrenia 1 |
333 |
0.89 |
| chr12_47613140_47613338 | 0.37 |
PCED1B |
PC-esterase domain containing 1B |
2858 |
0.3 |
| chrX_115456567_115456718 | 0.37 |
ENSG00000207033 |
. |
75471 |
0.11 |
| chr7_28369849_28370167 | 0.37 |
CREB5 |
cAMP responsive element binding protein 5 |
31068 |
0.23 |
| chr8_141599639_141599927 | 0.37 |
AGO2 |
argonaute RISC catalytic component 2 |
16204 |
0.22 |
| chr12_865415_865972 | 0.37 |
WNK1 |
WNK lysine deficient protein kinase 1 |
2961 |
0.28 |
| chr12_92524835_92525431 | 0.37 |
C12orf79 |
chromosome 12 open reading frame 79 |
5664 |
0.19 |
| chr20_52199257_52199767 | 0.37 |
ZNF217 |
zinc finger protein 217 |
124 |
0.96 |
| chr4_154855782_154855933 | 0.37 |
ENSG00000252181 |
. |
61254 |
0.14 |
| chr1_55701600_55701751 | 0.37 |
ENSG00000265822 |
. |
10361 |
0.22 |
| chr14_22977741_22978031 | 0.37 |
TRAJ15 |
T cell receptor alpha joining 15 |
20694 |
0.09 |
| chr5_130617107_130617383 | 0.36 |
CDC42SE2 |
CDC42 small effector 2 |
17452 |
0.27 |
| chr5_54450782_54450933 | 0.36 |
GPX8 |
glutathione peroxidase 8 (putative) |
5089 |
0.13 |
| chr13_103357618_103357769 | 0.36 |
METTL21C |
methyltransferase like 21C |
10839 |
0.14 |
| chr7_150146102_150146542 | 0.36 |
GIMAP8 |
GTPase, IMAP family member 8 |
1396 |
0.39 |
| chr1_89531148_89531299 | 0.36 |
GBP1 |
guanylate binding protein 1, interferon-inducible |
180 |
0.94 |
| chrY_15016828_15017148 | 0.36 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
246 |
0.96 |
| chr5_67576190_67576341 | 0.36 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
130 |
0.98 |
| chr14_22458708_22459003 | 0.36 |
ENSG00000238634 |
. |
152032 |
0.04 |
| chr21_32553132_32553283 | 0.35 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
50668 |
0.17 |
| chr9_124991302_124991453 | 0.35 |
LHX6 |
LIM homeobox 6 |
157 |
0.94 |
| chr16_80603632_80603783 | 0.35 |
RP11-525K10.3 |
|
6675 |
0.18 |
| chr16_11346250_11346518 | 0.35 |
RMI2 |
RecQ mediated genome instability 2 |
2878 |
0.13 |
| chr6_119398487_119399078 | 0.35 |
FAM184A |
family with sequence similarity 184, member A |
755 |
0.67 |
| chr6_36985827_36986032 | 0.35 |
FGD2 |
FYVE, RhoGEF and PH domain containing 2 |
12506 |
0.17 |
| chr6_160883573_160883724 | 0.35 |
LPAL2 |
lipoprotein, Lp(a)-like 2, pseudogene |
48391 |
0.17 |
| chr14_22982199_22982350 | 0.35 |
TRAJ15 |
T cell receptor alpha joining 15 |
16306 |
0.1 |
| chr16_86795095_86795257 | 0.35 |
FOXL1 |
forkhead box L1 |
183061 |
0.03 |
| chr1_26704591_26705236 | 0.35 |
ZNF683 |
zinc finger protein 683 |
3900 |
0.15 |
| chr1_151795357_151795508 | 0.35 |
RORC |
RAR-related orphan receptor C |
3135 |
0.1 |
| chr10_92079410_92079561 | 0.34 |
ENSG00000222451 |
. |
155768 |
0.04 |
| chr8_128811686_128811837 | 0.34 |
ENSG00000249859 |
. |
3553 |
0.33 |
| chr6_16421411_16421759 | 0.34 |
ENSG00000265642 |
. |
7169 |
0.31 |
| chr2_138710392_138710640 | 0.34 |
HNMT |
histamine N-methyltransferase |
11074 |
0.32 |
| chr3_5233234_5233385 | 0.34 |
EDEM1 |
ER degradation enhancer, mannosidase alpha-like 1 |
3275 |
0.19 |
| chr4_109082592_109082835 | 0.34 |
LEF1 |
lymphoid enhancer-binding factor 1 |
4744 |
0.25 |
| chr11_102189311_102189462 | 0.34 |
BIRC3 |
baculoviral IAP repeat containing 3 |
1092 |
0.48 |
| chr16_66907243_66907451 | 0.34 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
188 |
0.9 |
| chr2_196523306_196523815 | 0.33 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
1387 |
0.5 |
| chr10_52191698_52191849 | 0.33 |
AC069547.2 |
Uncharacterized protein |
39007 |
0.15 |
| chr17_43389731_43389977 | 0.33 |
ENSG00000199953 |
. |
15009 |
0.13 |
| chr22_40307152_40307436 | 0.33 |
GRAP2 |
GRB2-related adaptor protein 2 |
10181 |
0.17 |
| chrX_13680096_13680380 | 0.33 |
TCEANC |
transcription elongation factor A (SII) N-terminal and central domain containing |
6824 |
0.2 |
| chr8_23075544_23076103 | 0.33 |
ENSG00000246582 |
. |
6161 |
0.12 |
| chr10_134776485_134776636 | 0.32 |
TTC40 |
tetratricopeptide repeat domain 40 |
20233 |
0.23 |
| chr10_8099206_8099457 | 0.32 |
GATA3 |
GATA binding protein 3 |
2562 |
0.44 |
| chr10_90030880_90031226 | 0.32 |
ENSG00000200891 |
. |
276601 |
0.01 |
| chr6_209971_210123 | 0.32 |
DUSP22 |
dual specificity phosphatase 22 |
81583 |
0.11 |
| chr10_124135009_124135427 | 0.32 |
PLEKHA1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
868 |
0.6 |
| chr1_9030857_9031244 | 0.32 |
ENSG00000265141 |
. |
8883 |
0.16 |
| chr1_16292676_16293171 | 0.32 |
ZBTB17 |
zinc finger and BTB domain containing 17 |
9651 |
0.13 |
| chr18_67624277_67624428 | 0.32 |
CD226 |
CD226 molecule |
60 |
0.99 |
| chr15_61013207_61013362 | 0.32 |
ENSG00000212625 |
. |
15684 |
0.2 |
| chr9_14029932_14030102 | 0.32 |
NFIB |
nuclear factor I/B |
150774 |
0.05 |
| chr2_31639599_31639781 | 0.32 |
XDH |
xanthine dehydrogenase |
2109 |
0.44 |
| chr11_35163942_35164467 | 0.32 |
CD44 |
CD44 molecule (Indian blood group) |
3351 |
0.21 |
| chr8_42748827_42749465 | 0.32 |
ENSG00000266044 |
. |
2272 |
0.19 |
| chr1_78471059_78471251 | 0.32 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
617 |
0.53 |
| chr17_38019811_38019962 | 0.32 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
493 |
0.74 |
| chr17_73680522_73680673 | 0.31 |
RP11-474I11.7 |
|
282 |
0.82 |
| chr3_181342247_181342398 | 0.31 |
SOX2-OT |
SOX2 overlapping transcript (non-protein coding) |
13925 |
0.28 |
| chr9_128463330_128463481 | 0.31 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
6051 |
0.28 |
| chr21_32533248_32533535 | 0.31 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
30852 |
0.23 |
| chr8_135643662_135643813 | 0.31 |
ZFAT |
zinc finger and AT hook domain containing |
8386 |
0.27 |
| chr15_86247440_86247591 | 0.31 |
RP11-815J21.1 |
|
3175 |
0.16 |
| chr7_2910047_2910285 | 0.31 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
26208 |
0.2 |
| chr13_98859522_98859872 | 0.31 |
ENSG00000263399 |
. |
1081 |
0.44 |
| chr20_25290950_25291163 | 0.31 |
ABHD12 |
abhydrolase domain containing 12 |
922 |
0.6 |
| chr1_85763681_85764097 | 0.31 |
ENSG00000264380 |
. |
13596 |
0.14 |
| chr19_1168749_1168988 | 0.30 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
74 |
0.95 |
| chr9_33446136_33446606 | 0.30 |
AQP3 |
aquaporin 3 (Gill blood group) |
1238 |
0.41 |
| chr10_33405368_33405626 | 0.30 |
ENSG00000263576 |
. |
17933 |
0.22 |
| chr2_151335990_151336454 | 0.30 |
RND3 |
Rho family GTPase 3 |
5674 |
0.35 |
| chr2_231191950_231192650 | 0.30 |
SP140L |
SP140 nuclear body protein-like |
315 |
0.92 |
| chr12_33072283_33072434 | 0.30 |
PKP2 |
plakophilin 2 |
22584 |
0.26 |
| chr10_47640402_47640757 | 0.30 |
ANTXRLP1 |
anthrax toxin receptor-like pseudogene 1 |
4212 |
0.25 |
| chr18_60825953_60826104 | 0.30 |
RP11-299P2.1 |
|
7475 |
0.24 |
| chr2_12862265_12862722 | 0.30 |
TRIB2 |
tribbles pseudokinase 2 |
4129 |
0.3 |
| chr15_33493746_33494182 | 0.30 |
FMN1 |
formin 1 |
7067 |
0.29 |
| chr6_142948626_142948785 | 0.30 |
RP1-67K17.3 |
|
120875 |
0.06 |
| chr13_110999770_111000142 | 0.30 |
COL4A2 |
collagen, type IV, alpha 2 |
40342 |
0.15 |
| chr7_130783126_130783277 | 0.30 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
8588 |
0.21 |
| chr9_18268349_18268500 | 0.30 |
ADAMTSL1 |
ADAMTS-like 1 |
205468 |
0.03 |
| chr18_64911369_64911711 | 0.30 |
ENSG00000265580 |
. |
51979 |
0.18 |
| chr2_162915924_162916075 | 0.30 |
AC008063.2 |
|
13767 |
0.2 |
| chr6_33384140_33384291 | 0.30 |
PHF1 |
PHD finger protein 1 |
1373 |
0.23 |
| chr10_76345165_76345892 | 0.30 |
ENSG00000206756 |
. |
34378 |
0.2 |
| chr7_14022721_14022872 | 0.29 |
ETV1 |
ets variant 1 |
3270 |
0.34 |
| chrY_20488207_20488358 | 0.29 |
ENSG00000251953 |
. |
19842 |
0.19 |
| chr1_158902534_158902685 | 0.29 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
1251 |
0.51 |
| chrX_2553752_2553903 | 0.29 |
CD99P1 |
CD99 molecule pseudogene 1 |
26371 |
0.2 |
| chr2_235402543_235402910 | 0.29 |
ARL4C |
ADP-ribosylation factor-like 4C |
2518 |
0.45 |
| chr13_41236803_41237000 | 0.29 |
FOXO1 |
forkhead box O1 |
3833 |
0.29 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.2 | 0.6 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.2 | 0.5 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.1 | 0.1 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 0.3 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.3 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.4 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.1 | 0.3 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.3 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.3 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.1 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.2 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.1 | 0.2 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 | 0.2 | GO:0002913 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.2 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.3 | GO:0072698 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.1 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.2 | GO:1901985 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.1 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 0.3 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.1 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.4 | GO:0002423 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.1 | 0.3 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.4 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.2 | GO:0061117 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0014808 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 | 0.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.3 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.3 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.2 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.5 | GO:0042130 | negative regulation of T cell proliferation(GO:0042130) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0032351 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0051323 | mitotic metaphase(GO:0000089) metaphase(GO:0051323) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.0 | GO:0046621 | negative regulation of organ growth(GO:0046621) |
| 0.0 | 0.2 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.0 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.4 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.2 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.1 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0032691 | negative regulation of interleukin-1 beta production(GO:0032691) |
| 0.0 | 0.0 | GO:0051608 | histamine transport(GO:0051608) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.0 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.0 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.1 | GO:0090312 | regulation of protein deacetylation(GO:0090311) positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.4 | GO:0006997 | nucleus organization(GO:0006997) |
| 0.0 | 0.2 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0045061 | thymic T cell selection(GO:0045061) |
| 0.0 | 0.0 | GO:0002823 | negative regulation of adaptive immune response(GO:0002820) negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002823) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.0 | GO:0035268 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.1 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.0 | GO:0060087 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.0 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.2 | GO:0042308 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein localization to nucleus(GO:1900181) negative regulation of protein import(GO:1904590) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0090559 | regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:2000644 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.0 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:0071326 | cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.1 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.1 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.0 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.2 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.0 | GO:0060896 | neural plate pattern specification(GO:0060896) |
| 0.0 | 0.0 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.0 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0010171 | body morphogenesis(GO:0010171) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.5 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0031083 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.5 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.0 | GO:0044216 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.0 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.0 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.2 | 0.5 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.4 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.3 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.5 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.4 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.2 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.6 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.2 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |