Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
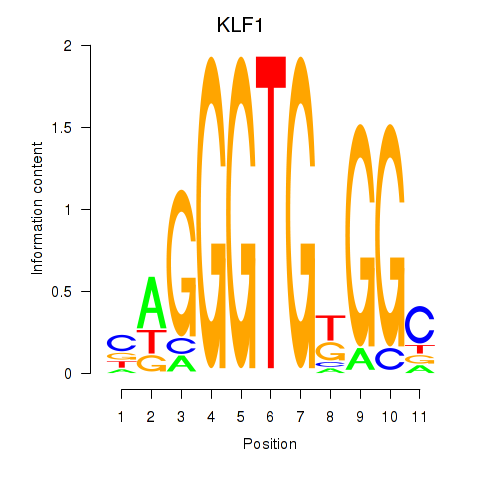
Results for KLF1
Z-value: 0.91
Transcription factors associated with KLF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF1
|
ENSG00000105610.4 | Kruppel like factor 1 |
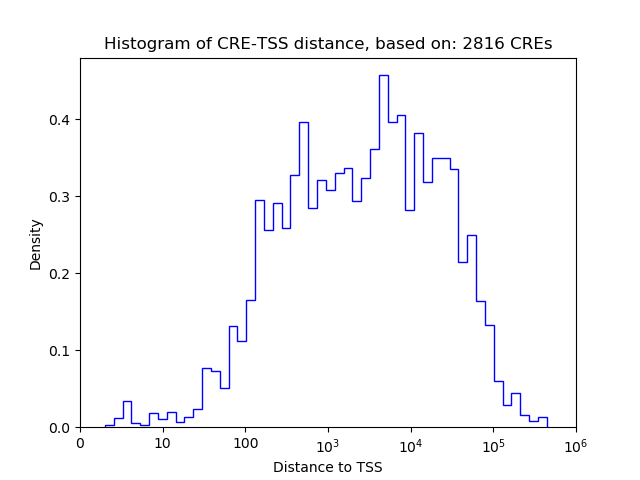
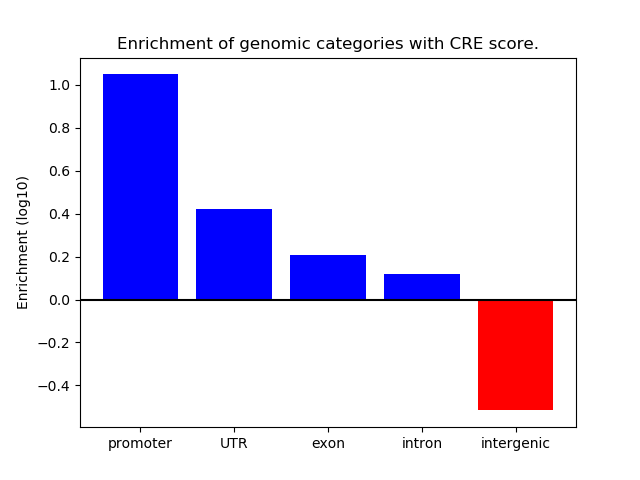
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_12997276_12997427 | KLF1 | 644 | 0.407609 | 0.61 | 8.0e-02 | Click! |
| chr19_12996769_12996974 | KLF1 | 1124 | 0.215101 | 0.60 | 8.8e-02 | Click! |
| chr19_12996052_12996768 | KLF1 | 1585 | 0.144282 | 0.29 | 4.5e-01 | Click! |
| chr19_12995815_12995996 | KLF1 | 2090 | 0.108974 | 0.26 | 5.1e-01 | Click! |
Activity of the KLF1 motif across conditions
Conditions sorted by the z-value of the KLF1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
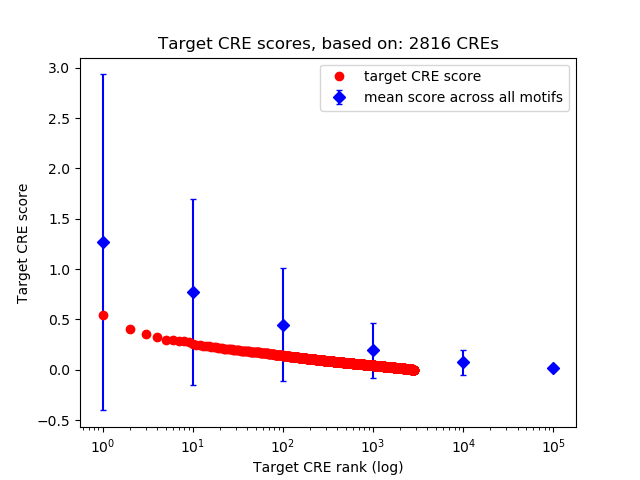
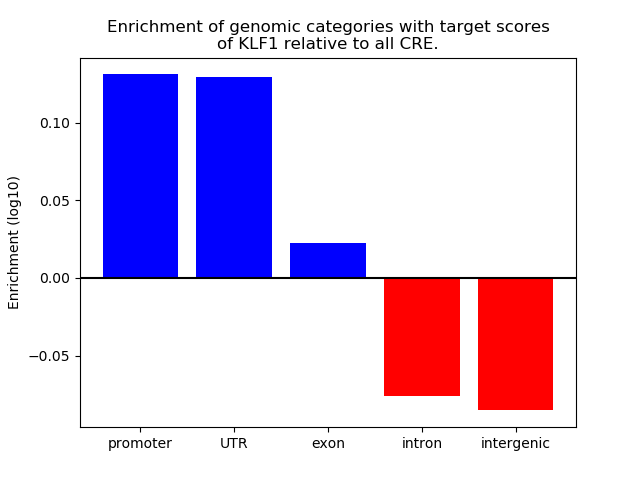
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_73718374_73718525 | 0.54 |
ITGB4 |
integrin, beta 4 |
872 |
0.43 |
| chr2_86055147_86055298 | 0.41 |
ENSG00000252321 |
. |
7487 |
0.17 |
| chr2_160918347_160919586 | 0.36 |
PLA2R1 |
phospholipase A2 receptor 1, 180kDa |
155 |
0.98 |
| chr18_33333968_33334119 | 0.32 |
GALNT1 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) |
99423 |
0.07 |
| chr8_25105948_25106099 | 0.30 |
DOCK5 |
dedicator of cytokinesis 5 |
50435 |
0.16 |
| chr10_126840512_126840747 | 0.29 |
CTBP2 |
C-terminal binding protein 2 |
6656 |
0.3 |
| chr1_956379_957022 | 0.29 |
AGRN |
agrin |
1197 |
0.27 |
| chr1_243651138_243651433 | 0.29 |
RP11-269F20.1 |
|
57549 |
0.15 |
| chr18_28958892_28959043 | 0.27 |
DSG4 |
desmoglein 4 |
2121 |
0.23 |
| chr1_183558268_183558419 | 0.26 |
NCF2 |
neutrophil cytosolic factor 2 |
1373 |
0.44 |
| chr17_7348839_7349178 | 0.25 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
158 |
0.84 |
| chr22_45635380_45635531 | 0.25 |
KIAA0930 |
KIAA0930 |
1195 |
0.46 |
| chr3_133614813_133615159 | 0.24 |
RAB6B |
RAB6B, member RAS oncogene family |
306 |
0.92 |
| chr11_130029601_130029752 | 0.24 |
ST14 |
suppression of tumorigenicity 14 (colon carcinoma) |
219 |
0.96 |
| chr2_113735443_113735594 | 0.24 |
IL36G |
interleukin 36, gamma |
78 |
0.97 |
| chr22_19102804_19102955 | 0.23 |
AC004471.9 |
|
6163 |
0.12 |
| chr7_150755256_150755900 | 0.23 |
CDK5 |
cyclin-dependent kinase 5 |
39 |
0.77 |
| chr10_82245695_82245846 | 0.22 |
TSPAN14 |
tetraspanin 14 |
26712 |
0.17 |
| chr3_184209968_184210530 | 0.22 |
EIF2B5-IT1 |
EIF2B5 intronic transcript 1 (non-protein coding) |
12652 |
0.18 |
| chr10_50398410_50398651 | 0.22 |
C10orf128 |
chromosome 10 open reading frame 128 |
2094 |
0.33 |
| chr17_76123581_76124282 | 0.21 |
TMC6 |
transmembrane channel-like 6 |
780 |
0.49 |
| chr11_65346255_65346406 | 0.20 |
EHBP1L1 |
EH domain binding protein 1-like 1 |
2813 |
0.1 |
| chr13_99328229_99328380 | 0.20 |
ENSG00000243366 |
. |
4070 |
0.29 |
| chr3_49943448_49943849 | 0.20 |
CTD-2330K9.3 |
Uncharacterized protein |
2228 |
0.16 |
| chr9_113979_114607 | 0.20 |
FOXD4 |
forkhead box D4 |
4124 |
0.25 |
| chr17_16977527_16977700 | 0.20 |
MPRIP-AS1 |
MPRIP antisense RNA 1 |
3453 |
0.23 |
| chr19_13113568_13113719 | 0.20 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
6991 |
0.09 |
| chr12_114334146_114334297 | 0.20 |
RP11-780K2.1 |
|
36153 |
0.21 |
| chr7_158750572_158750973 | 0.20 |
ENSG00000231419 |
. |
50340 |
0.16 |
| chr22_38291318_38291469 | 0.19 |
ENSG00000207227 |
. |
4039 |
0.12 |
| chr8_22847583_22847734 | 0.19 |
RHOBTB2 |
Rho-related BTB domain containing 2 |
2663 |
0.19 |
| chr10_120739922_120740073 | 0.19 |
RP11-498J9.2 |
|
23399 |
0.15 |
| chr8_144657322_144657645 | 0.19 |
RP11-661A12.9 |
|
1823 |
0.12 |
| chr3_49170609_49171370 | 0.19 |
LAMB2 |
laminin, beta 2 (laminin S) |
438 |
0.69 |
| chr8_144697320_144697471 | 0.19 |
TSTA3 |
tissue specific transplantation antigen P35B |
1239 |
0.21 |
| chr10_123313697_123313848 | 0.19 |
FGFR2 |
fibroblast growth factor receptor 2 |
22944 |
0.27 |
| chr2_114260314_114260465 | 0.19 |
FOXD4L1 |
forkhead box D4-like 1 |
3728 |
0.22 |
| chr10_46010179_46010570 | 0.19 |
MARCH8 |
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
20445 |
0.22 |
| chr14_24783731_24783882 | 0.19 |
LTB4R |
leukotriene B4 receptor |
100 |
0.9 |
| chr15_77286572_77286762 | 0.18 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
759 |
0.66 |
| chr10_74082676_74082827 | 0.18 |
DNAJB12 |
DnaJ (Hsp40) homolog, subfamily B, member 12 |
21968 |
0.16 |
| chr22_23599471_23599622 | 0.18 |
BCR |
breakpoint cluster region |
3399 |
0.21 |
| chr19_12274174_12274895 | 0.18 |
ZNF136 |
zinc finger protein 136 |
603 |
0.62 |
| chr1_958272_958423 | 0.18 |
AGRN |
agrin |
2844 |
0.12 |
| chr18_76261468_76261619 | 0.18 |
ENSG00000201723 |
. |
42645 |
0.22 |
| chr16_50313127_50313376 | 0.18 |
ADCY7 |
adenylate cyclase 7 |
187 |
0.95 |
| chr2_114083135_114083388 | 0.18 |
PAX8 |
paired box 8 |
46734 |
0.11 |
| chr17_73860377_73860686 | 0.18 |
WBP2 |
WW domain binding protein 2 |
7943 |
0.08 |
| chr5_6767017_6767234 | 0.17 |
PAPD7 |
PAP associated domain containing 7 |
52380 |
0.15 |
| chr18_46460407_46461027 | 0.17 |
SMAD7 |
SMAD family member 7 |
14158 |
0.24 |
| chr17_43248472_43248682 | 0.17 |
RP13-890H12.2 |
|
275 |
0.83 |
| chr9_132406657_132406808 | 0.17 |
ASB6 |
ankyrin repeat and SOCS box containing 6 |
2288 |
0.19 |
| chr20_39970036_39970187 | 0.17 |
LPIN3 |
lipin 3 |
551 |
0.77 |
| chr9_137275051_137275202 | 0.17 |
ENSG00000263897 |
. |
3869 |
0.28 |
| chr17_35214294_35214445 | 0.17 |
RP11-445F12.1 |
|
79552 |
0.07 |
| chr8_145009573_145009909 | 0.17 |
PLEC |
plectin |
4017 |
0.13 |
| chr10_129843979_129844130 | 0.17 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
1616 |
0.51 |
| chr15_41195326_41195612 | 0.17 |
VPS18 |
vacuolar protein sorting 18 homolog (S. cerevisiae) |
8804 |
0.11 |
| chr11_3181150_3181578 | 0.17 |
OSBPL5 |
oxysterol binding protein-like 5 |
1259 |
0.4 |
| chr9_137248904_137249055 | 0.17 |
ENSG00000263897 |
. |
22278 |
0.21 |
| chr21_45590544_45590695 | 0.17 |
AP001055.1 |
|
2961 |
0.19 |
| chr17_48998464_48998615 | 0.17 |
TOB1 |
transducer of ERBB2, 1 |
53200 |
0.1 |
| chr14_96691207_96691358 | 0.17 |
RP11-404P21.1 |
|
9156 |
0.12 |
| chr11_12112400_12112551 | 0.17 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
3068 |
0.36 |
| chr3_48636571_48636823 | 0.16 |
COL7A1 |
collagen, type VII, alpha 1 |
3997 |
0.11 |
| chr1_155102531_155102783 | 0.16 |
EFNA1 |
ephrin-A1 |
2305 |
0.11 |
| chr2_134898080_134898231 | 0.16 |
ENSG00000263813 |
. |
13459 |
0.23 |
| chr16_15236302_15236453 | 0.16 |
ENSG00000207294 |
. |
8177 |
0.11 |
| chr16_31213085_31213654 | 0.16 |
C16orf98 |
chromosome 16 open reading frame 98 |
163 |
0.81 |
| chr18_3327008_3327159 | 0.16 |
MYL12B |
myosin, light chain 12B, regulatory |
64129 |
0.09 |
| chr20_60388194_60388345 | 0.16 |
RP11-429E11.2 |
|
58157 |
0.14 |
| chr9_137444566_137444717 | 0.16 |
COL5A1 |
collagen, type V, alpha 1 |
88979 |
0.08 |
| chr1_38510289_38510696 | 0.16 |
POU3F1 |
POU class 3 homeobox 1 |
1958 |
0.27 |
| chr14_105436556_105437076 | 0.16 |
AHNAK2 |
AHNAK nucleoprotein 2 |
7878 |
0.16 |
| chr17_76879212_76879961 | 0.16 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
9354 |
0.14 |
| chrX_153666039_153666841 | 0.16 |
GDI1 |
GDP dissociation inhibitor 1 |
1174 |
0.23 |
| chr14_100048320_100048471 | 0.16 |
ENSG00000200506 |
. |
959 |
0.49 |
| chr15_29434000_29434151 | 0.16 |
ENSG00000252868 |
. |
91997 |
0.08 |
| chr20_17502066_17502217 | 0.15 |
BFSP1 |
beaded filament structural protein 1, filensin |
9873 |
0.23 |
| chr19_49840027_49840178 | 0.15 |
CD37 |
CD37 molecule |
370 |
0.72 |
| chr1_3390049_3390200 | 0.15 |
ARHGEF16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
1936 |
0.35 |
| chr6_108972255_108972979 | 0.15 |
FOXO3 |
forkhead box O3 |
4932 |
0.33 |
| chr1_156877457_156877608 | 0.15 |
LRRC71 |
leucine rich repeat containing 71 |
12910 |
0.12 |
| chr8_23754300_23754592 | 0.15 |
STC1 |
stanniocalcin 1 |
42126 |
0.17 |
| chr7_116141197_116141631 | 0.15 |
CAV2 |
caveolin 2 |
1580 |
0.29 |
| chr6_36684370_36684521 | 0.15 |
RAB44 |
RAB44, member RAS oncogene family |
1189 |
0.44 |
| chrX_49033222_49033373 | 0.15 |
PLP2 |
proteolipid protein 2 (colonic epithelium-enriched) |
5024 |
0.09 |
| chr7_75357800_75357951 | 0.15 |
HIP1 |
huntingtin interacting protein 1 |
10384 |
0.21 |
| chr19_3097024_3097175 | 0.15 |
GNA11 |
guanine nucleotide binding protein (G protein), alpha 11 (Gq class) |
2691 |
0.16 |
| chr12_122468241_122468392 | 0.15 |
BCL7A |
B-cell CLL/lymphoma 7A |
8524 |
0.22 |
| chr17_46103279_46103479 | 0.14 |
COPZ2 |
coatomer protein complex, subunit zeta 2 |
7859 |
0.09 |
| chr9_135819104_135820215 | 0.14 |
TSC1 |
tuberous sclerosis 1 |
349 |
0.7 |
| chr10_45476062_45476312 | 0.14 |
C10orf10 |
chromosome 10 open reading frame 10 |
1929 |
0.22 |
| chr22_39147328_39147855 | 0.14 |
SUN2 |
Sad1 and UNC84 domain containing 2 |
1272 |
0.23 |
| chr2_75185375_75185738 | 0.14 |
POLE4 |
polymerase (DNA-directed), epsilon 4, accessory subunit |
63 |
0.98 |
| chr4_25031967_25032301 | 0.14 |
LGI2 |
leucine-rich repeat LGI family, member 2 |
153 |
0.97 |
| chr11_64951074_64951225 | 0.14 |
CAPN1 |
calpain 1, (mu/I) large subunit |
342 |
0.74 |
| chr7_87257054_87258255 | 0.14 |
RUNDC3B |
RUN domain containing 3B |
75 |
0.97 |
| chr9_14322549_14323229 | 0.14 |
NFIB |
nuclear factor I/B |
552 |
0.82 |
| chr8_144714709_144714860 | 0.14 |
RP11-661A12.12 |
|
1142 |
0.24 |
| chr9_114361613_114362271 | 0.14 |
PTGR1 |
prostaglandin reductase 1 |
33 |
0.97 |
| chr15_96903147_96903373 | 0.14 |
AC087477.1 |
Uncharacterized protein |
1227 |
0.45 |
| chr3_5055062_5055354 | 0.14 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
33562 |
0.15 |
| chr18_74779548_74779839 | 0.14 |
MBP |
myelin basic protein |
37524 |
0.2 |
| chrX_41301675_41302169 | 0.14 |
NYX |
nyctalopin |
4765 |
0.25 |
| chr11_72451789_72452038 | 0.14 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
11535 |
0.12 |
| chr9_137235953_137236130 | 0.14 |
RXRA |
retinoid X receptor, alpha |
17615 |
0.22 |
| chr14_24784022_24784248 | 0.14 |
LTB4R |
leukotriene B4 receptor |
229 |
0.8 |
| chr12_120662827_120663239 | 0.14 |
PXN |
paxillin |
480 |
0.71 |
| chr11_1897109_1897260 | 0.14 |
LSP1 |
lymphocyte-specific protein 1 |
523 |
0.63 |
| chr16_389783_389934 | 0.14 |
AXIN1 |
axin 1 |
12591 |
0.1 |
| chr15_47476270_47477144 | 0.14 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
409 |
0.87 |
| chr22_46475652_46475803 | 0.13 |
FLJ27365 |
hsa-mir-4763 |
465 |
0.67 |
| chr12_41582969_41583319 | 0.13 |
PDZRN4 |
PDZ domain containing ring finger 4 |
894 |
0.75 |
| chr17_75311931_75312082 | 0.13 |
SEPT9 |
septin 9 |
3591 |
0.27 |
| chr19_51433227_51433378 | 0.13 |
KLK4 |
kallikrein-related peptidase 4 |
19308 |
0.07 |
| chr17_43205399_43205692 | 0.13 |
PLCD3 |
phospholipase C, delta 3 |
4346 |
0.12 |
| chr5_68710635_68711790 | 0.13 |
MARVELD2 |
MARVEL domain containing 2 |
3 |
0.97 |
| chr19_2045769_2045920 | 0.13 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
3212 |
0.14 |
| chr2_106886104_106886958 | 0.13 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
75736 |
0.1 |
| chr5_111755303_111755604 | 0.13 |
EPB41L4A-AS2 |
EPB41L4A antisense RNA 2 (head to head) |
173 |
0.78 |
| chr19_11805219_11805471 | 0.13 |
CTC-499B15.4 |
|
11984 |
0.13 |
| chr2_69534033_69534297 | 0.13 |
ENSG00000252250 |
. |
27567 |
0.2 |
| chr6_125203961_125204112 | 0.13 |
RNF217 |
ring finger protein 217 |
79655 |
0.11 |
| chr2_43333094_43333245 | 0.13 |
ENSG00000207087 |
. |
14537 |
0.28 |
| chr11_64479921_64480072 | 0.13 |
NRXN2 |
neurexin 2 |
221 |
0.91 |
| chr7_5638418_5638613 | 0.13 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
4198 |
0.19 |
| chr2_28600742_28600893 | 0.13 |
FOSL2 |
FOS-like antigen 2 |
14852 |
0.16 |
| chr9_130590515_130590893 | 0.13 |
ENSG00000222421 |
. |
4318 |
0.09 |
| chr7_5638779_5639073 | 0.13 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
4609 |
0.19 |
| chr1_43473710_43473861 | 0.13 |
ENSG00000252803 |
. |
15523 |
0.17 |
| chr4_1288964_1289115 | 0.13 |
MAEA |
macrophage erythroblast attacher |
5356 |
0.16 |
| chr18_77664931_77665082 | 0.13 |
KCNG2 |
potassium voltage-gated channel, subfamily G, member 2 |
41338 |
0.14 |
| chr1_154979820_154980060 | 0.13 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
4645 |
0.08 |
| chr1_222884853_222885497 | 0.13 |
AIDA |
axin interactor, dorsalization associated |
595 |
0.46 |
| chr19_18201957_18202170 | 0.12 |
IL12RB1 |
interleukin 12 receptor, beta 1 |
4250 |
0.16 |
| chr19_5903751_5904369 | 0.12 |
FUT5 |
fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
262 |
0.34 |
| chr6_56614643_56614951 | 0.12 |
DST |
dystonin |
35880 |
0.21 |
| chr1_1280074_1280225 | 0.12 |
DVL1 |
dishevelled segment polarity protein 1 |
4343 |
0.08 |
| chr5_175199924_175200075 | 0.12 |
CPLX2 |
complexin 2 |
23314 |
0.2 |
| chr11_95770968_95771151 | 0.12 |
MTMR2 |
myotubularin related protein 2 |
113600 |
0.07 |
| chr19_1856293_1856444 | 0.12 |
CTB-31O20.8 |
|
3881 |
0.1 |
| chr10_24754741_24755189 | 0.12 |
KIAA1217 |
KIAA1217 |
495 |
0.84 |
| chr18_32990029_32990180 | 0.12 |
ZNF396 |
zinc finger protein 396 |
32803 |
0.17 |
| chr5_131795063_131795214 | 0.12 |
ENSG00000202533 |
. |
8701 |
0.13 |
| chr17_79989895_79990665 | 0.12 |
RAC3 |
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
220 |
0.81 |
| chr19_751908_752059 | 0.12 |
MISP |
mitotic spindle positioning |
857 |
0.44 |
| chr9_88358324_88358790 | 0.12 |
AGTPBP1 |
ATP/GTP binding protein 1 |
1613 |
0.56 |
| chr9_140089237_140089388 | 0.12 |
TPRN |
taperin |
5245 |
0.06 |
| chr7_1164684_1164835 | 0.12 |
C7orf50 |
chromosome 7 open reading frame 50 |
13117 |
0.12 |
| chr21_31311288_31312406 | 0.12 |
GRIK1 |
glutamate receptor, ionotropic, kainate 1 |
3 |
0.99 |
| chr20_61463695_61464080 | 0.12 |
COL9A3 |
collagen, type IX, alpha 3 |
14724 |
0.11 |
| chr3_186079383_186079733 | 0.12 |
DGKG |
diacylglycerol kinase, gamma 90kDa |
188 |
0.97 |
| chr15_81426568_81426818 | 0.12 |
C15orf26 |
chromosome 15 open reading frame 26 |
105 |
0.98 |
| chr19_16994486_16994637 | 0.12 |
F2RL3 |
coagulation factor II (thrombin) receptor-like 3 |
5110 |
0.13 |
| chr8_144653425_144654110 | 0.12 |
MROH6 |
maestro heat-like repeat family member 6 |
1151 |
0.21 |
| chr11_561033_561967 | 0.12 |
RASSF7 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
47 |
0.89 |
| chr1_110772672_110772823 | 0.12 |
KCNC4 |
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
18625 |
0.12 |
| chr15_41795749_41795900 | 0.12 |
ITPKA |
inositol-trisphosphate 3-kinase A |
9751 |
0.13 |
| chr1_119543496_119543659 | 0.12 |
TBX15 |
T-box 15 |
11398 |
0.25 |
| chr2_97198141_97198292 | 0.12 |
RP11-363D14.1 |
|
173 |
0.95 |
| chr3_136677917_136678068 | 0.12 |
IL20RB |
interleukin 20 receptor beta |
1141 |
0.51 |
| chr20_62710337_62711142 | 0.12 |
RGS19 |
regulator of G-protein signaling 19 |
106 |
0.88 |
| chr9_22006120_22006693 | 0.12 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
2546 |
0.22 |
| chr5_150159569_150159738 | 0.12 |
AC010441.1 |
|
1786 |
0.26 |
| chr13_50194281_50194827 | 0.12 |
ARL11 |
ADP-ribosylation factor-like 11 |
7881 |
0.21 |
| chr3_53033553_53033704 | 0.12 |
SFMBT1 |
Scm-like with four mbt domains 1 |
45653 |
0.12 |
| chr7_101796468_101796619 | 0.12 |
ENSG00000252824 |
. |
40901 |
0.15 |
| chr9_137248251_137248693 | 0.12 |
ENSG00000263897 |
. |
22785 |
0.21 |
| chr9_137258318_137258525 | 0.12 |
ENSG00000263897 |
. |
12836 |
0.24 |
| chr1_209603784_209603935 | 0.12 |
ENSG00000230937 |
. |
1619 |
0.52 |
| chr6_7553448_7553719 | 0.11 |
DSP |
desmoplakin |
11733 |
0.19 |
| chr3_49384336_49384565 | 0.11 |
USP4 |
ubiquitin specific peptidase 4 (proto-oncogene) |
6946 |
0.11 |
| chr14_61123462_61124719 | 0.11 |
SIX1 |
SIX homeobox 1 |
887 |
0.64 |
| chr2_217501949_217502100 | 0.11 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
3906 |
0.22 |
| chr17_70515597_70515748 | 0.11 |
ENSG00000200783 |
. |
144619 |
0.05 |
| chr17_7342306_7342601 | 0.11 |
FGF11 |
fibroblast growth factor 11 |
107 |
0.87 |
| chr20_55754754_55754905 | 0.11 |
RP4-813D12.2 |
|
35099 |
0.17 |
| chr18_55746256_55746407 | 0.11 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
31873 |
0.2 |
| chr17_64670217_64670368 | 0.11 |
AC006947.1 |
|
2198 |
0.33 |
| chr14_77512028_77512179 | 0.11 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
17069 |
0.17 |
| chr17_18280756_18280929 | 0.11 |
EVPLL |
envoplakin-like |
134 |
0.93 |
| chr9_140130864_140131015 | 0.11 |
TUBB4B |
tubulin, beta 4B class IVb |
4726 |
0.06 |
| chr17_76136415_76136566 | 0.11 |
C17orf99 |
chromosome 17 open reading frame 99 |
5944 |
0.12 |
| chr1_227015373_227015524 | 0.11 |
PSEN2 |
presenilin 2 (Alzheimer disease 4) |
42437 |
0.16 |
| chr5_10635837_10635988 | 0.11 |
ANKRD33B-AS1 |
ANKRD33B antisense RNA 1 |
7575 |
0.25 |
| chr1_40425789_40425948 | 0.11 |
MFSD2A |
major facilitator superfamily domain containing 2A |
5046 |
0.17 |
| chr15_31653592_31653811 | 0.11 |
KLF13 |
Kruppel-like factor 13 |
4656 |
0.34 |
| chr3_12700084_12700501 | 0.11 |
RAF1 |
v-raf-1 murine leukemia viral oncogene homolog 1 |
5324 |
0.21 |
| chr19_41933992_41934158 | 0.11 |
B3GNT8 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
560 |
0.56 |
| chr6_10383738_10384217 | 0.11 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
20850 |
0.18 |
| chr10_134402361_134402743 | 0.11 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
18878 |
0.23 |
| chr4_185723588_185724132 | 0.11 |
ACSL1 |
acyl-CoA synthetase long-chain family member 1 |
3082 |
0.25 |
| chr7_121436163_121436314 | 0.11 |
ENSG00000252704 |
. |
59741 |
0.14 |
| chrX_22670943_22671094 | 0.11 |
ENSG00000265819 |
. |
15909 |
0.29 |
| chr16_67213263_67213414 | 0.11 |
KIAA0895L |
KIAA0895-like |
627 |
0.43 |
| chr1_1244917_1245068 | 0.11 |
PUSL1 |
pseudouridylate synthase-like 1 |
791 |
0.33 |
| chr10_77167336_77167780 | 0.11 |
ENSG00000237149 |
. |
4746 |
0.21 |
| chr1_208374584_208374735 | 0.11 |
PLXNA2 |
plexin A2 |
43006 |
0.22 |
| chr16_2173775_2174214 | 0.11 |
RP11-304L19.2 |
|
4786 |
0.06 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.2 | GO:0003179 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0052167 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0032823 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0036296 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.2 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0036473 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0060678 | dichotomous subdivision of terminal units involved in ureteric bud branching(GO:0060678) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.0 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.1 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.0 | 0.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.0 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.0 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |