Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for KLF13
Z-value: 0.69
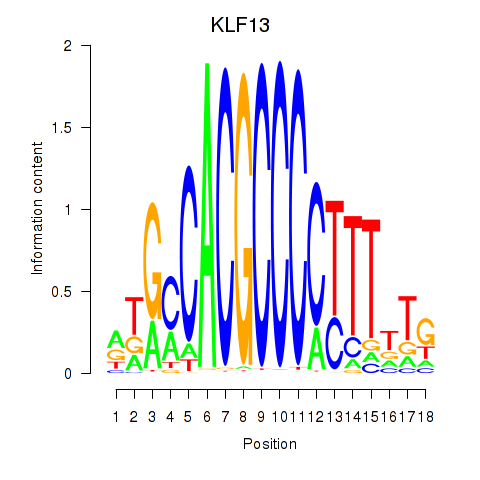
Transcription factors associated with KLF13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF13
|
ENSG00000169926.5 | Kruppel like factor 13 |
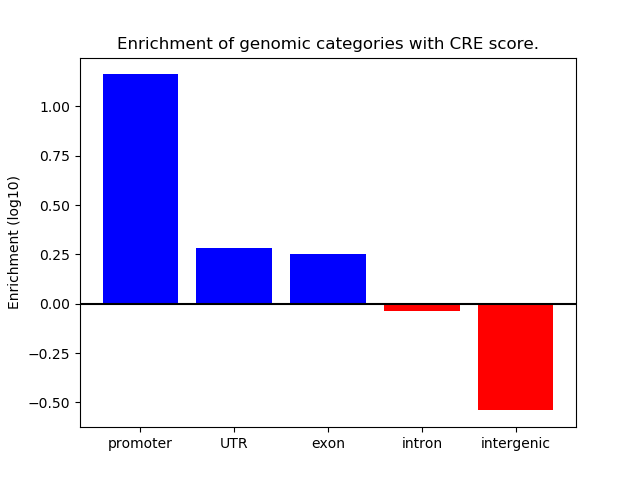
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_31732761_31733091 | KLF13 | 74569 | 0.120897 | -0.91 | 7.6e-04 | Click! |
| chr15_31631556_31631837 | KLF13 | 440 | 0.902680 | -0.82 | 6.6e-03 | Click! |
| chr15_31637211_31637362 | KLF13 | 5150 | 0.331718 | -0.81 | 8.7e-03 | Click! |
| chr15_31684023_31684332 | KLF13 | 25820 | 0.259140 | -0.76 | 1.9e-02 | Click! |
| chr15_31637569_31637720 | KLF13 | 5508 | 0.327395 | -0.75 | 2.1e-02 | Click! |
Activity of the KLF13 motif across conditions
Conditions sorted by the z-value of the KLF13 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
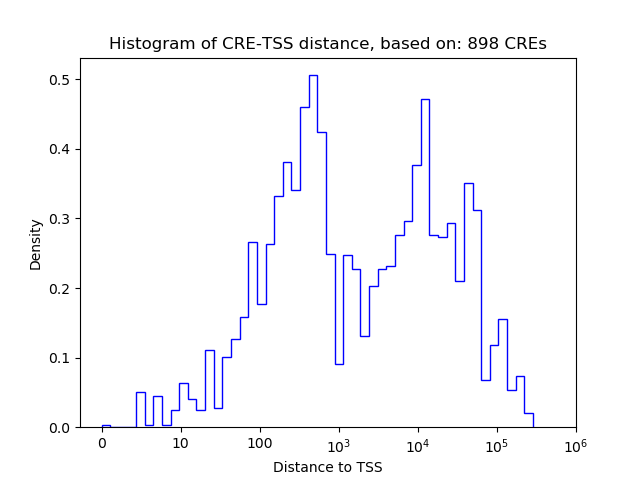
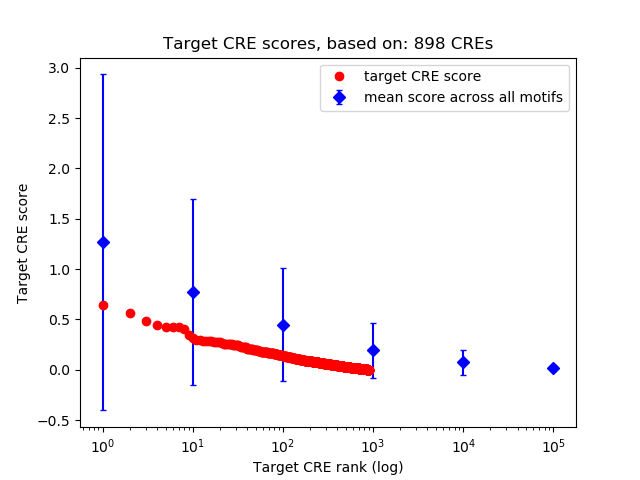
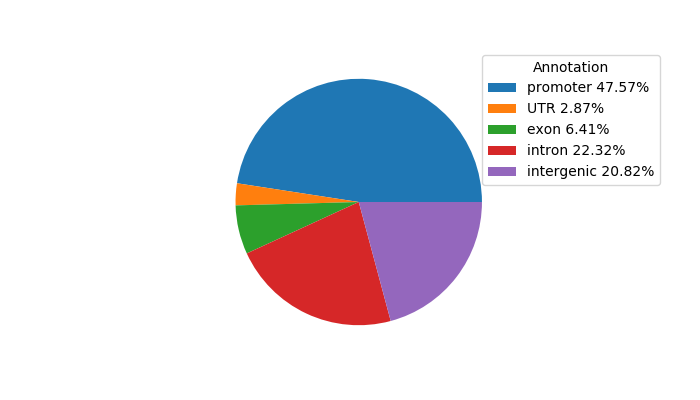
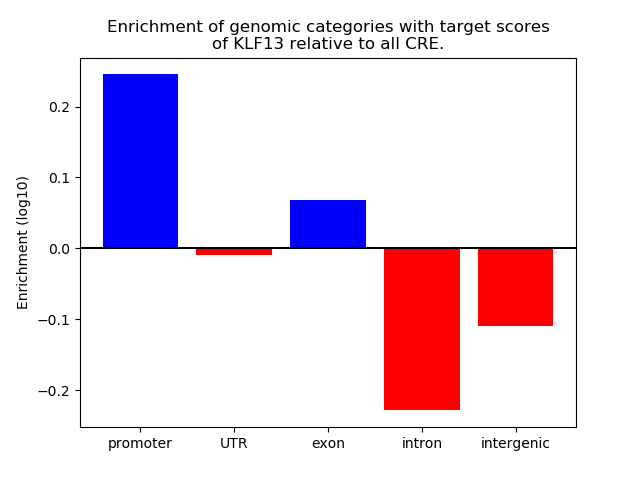
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_49866685_49866836 | 0.64 |
DKKL1 |
dickkopf-like 1 |
91 |
0.9 |
| chr17_26554461_26555359 | 0.56 |
PYY2 |
peptide YY, 2 (pseudogene) |
578 |
0.68 |
| chr1_3816010_3816785 | 0.48 |
C1orf174 |
chromosome 1 open reading frame 174 |
452 |
0.8 |
| chr12_1613703_1614089 | 0.44 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
25161 |
0.21 |
| chr17_13505362_13505866 | 0.43 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
370 |
0.91 |
| chr19_52452059_52452669 | 0.42 |
HCCAT3 |
hepatocellular carcinoma associated transcript 3 (non-protein coding) |
23 |
0.96 |
| chr19_3606102_3607092 | 0.42 |
TBXA2R |
thromboxane A2 receptor |
61 |
0.89 |
| chr1_15573540_15574233 | 0.41 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
110 |
0.97 |
| chr19_6740138_6741070 | 0.35 |
TRIP10 |
thyroid hormone receptor interactor 10 |
325 |
0.81 |
| chr5_133861629_133862874 | 0.31 |
JADE2 |
jade family PHD finger 2 |
76 |
0.97 |
| chr17_10102219_10102533 | 0.30 |
GAS7 |
growth arrest-specific 7 |
508 |
0.83 |
| chr10_50009233_50009384 | 0.29 |
WDFY4 |
WDFY family member 4 |
1475 |
0.49 |
| chr19_51161641_51162497 | 0.29 |
C19orf81 |
chromosome 19 open reading frame 81 |
9024 |
0.12 |
| chr6_159239962_159240825 | 0.28 |
EZR |
ezrin |
51 |
0.96 |
| chr21_32591500_32591929 | 0.28 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
57369 |
0.16 |
| chr4_187025490_187026623 | 0.28 |
FAM149A |
family with sequence similarity 149, member A |
175 |
0.94 |
| chr15_41055526_41056933 | 0.28 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
11 |
0.96 |
| chr1_25393209_25393360 | 0.28 |
ENSG00000264371 |
. |
43290 |
0.16 |
| chr11_108107293_108107444 | 0.28 |
ENSG00000206967 |
. |
7037 |
0.16 |
| chr5_140797768_140798225 | 0.27 |
PCDHGB7 |
protocadherin gamma subfamily B, 7 |
569 |
0.45 |
| chrX_129657730_129658601 | 0.27 |
RBMX2 |
RNA binding motif protein, X-linked 2 |
122186 |
0.05 |
| chrX_48910717_48911936 | 0.26 |
CCDC120 |
coiled-coil domain containing 120 |
225 |
0.85 |
| chr18_58201323_58201474 | 0.26 |
ENSG00000253011 |
. |
20978 |
0.26 |
| chr17_62705097_62705248 | 0.26 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
46986 |
0.12 |
| chr19_49866897_49867069 | 0.26 |
DKKL1 |
dickkopf-like 1 |
4 |
0.93 |
| chr9_125830879_125831036 | 0.25 |
GPR21 |
G protein-coupled receptor 21 |
34151 |
0.13 |
| chr1_7130330_7130574 | 0.25 |
RP11-334N17.1 |
|
56113 |
0.15 |
| chr9_97149391_97149542 | 0.25 |
HIATL1 |
hippocampus abundant transcript-like 1 |
12633 |
0.22 |
| chr1_221273332_221273483 | 0.25 |
HLX |
H2.0-like homeobox |
218823 |
0.02 |
| chr4_4388085_4389202 | 0.24 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
189 |
0.96 |
| chr13_33759866_33760149 | 0.24 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
209 |
0.95 |
| chr14_93313876_93314027 | 0.24 |
GOLGA5 |
golgin A5 |
14480 |
0.25 |
| chr10_94581983_94582410 | 0.24 |
EXOC6 |
exocyst complex component 6 |
8739 |
0.28 |
| chr2_121419142_121419293 | 0.23 |
ENSG00000201006 |
. |
10368 |
0.28 |
| chr19_10947512_10947663 | 0.23 |
C19orf38 |
chromosome 19 open reading frame 38 |
336 |
0.63 |
| chr11_74442538_74442948 | 0.23 |
CHRDL2 |
chordin-like 2 |
313 |
0.84 |
| chr21_34674464_34674615 | 0.23 |
IFNAR1 |
interferon (alpha, beta and omega) receptor 1 |
22195 |
0.14 |
| chr5_171604379_171604531 | 0.23 |
STK10 |
serine/threonine kinase 10 |
10935 |
0.19 |
| chr1_21934477_21934628 | 0.22 |
RAP1GAP |
RAP1 GTPase activating protein |
11982 |
0.16 |
| chr1_2210669_2210850 | 0.21 |
RP4-713A8.1 |
|
47373 |
0.08 |
| chr10_77168132_77169350 | 0.21 |
ENSG00000237149 |
. |
5929 |
0.2 |
| chr4_141077908_141078102 | 0.21 |
MAML3 |
mastermind-like 3 (Drosophila) |
2667 |
0.32 |
| chr18_25755743_25755974 | 0.21 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
1552 |
0.58 |
| chr6_54912113_54912264 | 0.21 |
RP3-523K23.2 |
|
104223 |
0.07 |
| chr17_76320400_76320721 | 0.20 |
SOCS3 |
suppressor of cytokine signaling 3 |
35595 |
0.11 |
| chr5_129241672_129241829 | 0.20 |
CTC-575N7.1 |
|
140 |
0.95 |
| chr1_225670324_225670475 | 0.20 |
RP11-496N12.6 |
|
17354 |
0.21 |
| chr16_83987226_83987552 | 0.20 |
OSGIN1 |
oxidative stress induced growth inhibitor 1 |
545 |
0.72 |
| chr19_3572978_3573964 | 0.20 |
MFSD12 |
major facilitator superfamily domain containing 12 |
156 |
0.77 |
| chr2_740358_740890 | 0.20 |
AC092159.3 |
|
47389 |
0.15 |
| chr19_55864944_55865497 | 0.19 |
COX6B2 |
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
688 |
0.4 |
| chr14_75744052_75744985 | 0.19 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
959 |
0.51 |
| chr17_60858241_60858392 | 0.19 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
20964 |
0.21 |
| chr8_105373799_105374058 | 0.19 |
DCSTAMP |
dendrocyte expressed seven transmembrane protein |
13159 |
0.2 |
| chr15_65023363_65023598 | 0.18 |
OAZ2 |
ornithine decarboxylase antizyme 2 |
28000 |
0.09 |
| chr8_9413349_9414633 | 0.18 |
RP11-375N15.2 |
|
39 |
0.92 |
| chr1_1118171_1118322 | 0.18 |
TTLL10 |
tubulin tyrosine ligase-like family, member 10 |
3169 |
0.1 |
| chr16_11406523_11406823 | 0.18 |
ENSG00000199668 |
. |
2251 |
0.14 |
| chr9_132369899_132370088 | 0.18 |
NTMT1 |
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
1170 |
0.36 |
| chr22_45125363_45125514 | 0.18 |
PRR5 |
proline rich 5 (renal) |
1246 |
0.47 |
| chr19_43968013_43968906 | 0.18 |
LYPD3 |
LY6/PLAUR domain containing 3 |
1353 |
0.34 |
| chr2_178937534_178937941 | 0.18 |
PDE11A |
phosphodiesterase 11A |
255 |
0.92 |
| chr17_41850254_41850405 | 0.17 |
DUSP3 |
dual specificity phosphatase 3 |
1807 |
0.22 |
| chr1_159823893_159825316 | 0.17 |
C1orf204 |
chromosome 1 open reading frame 204 |
533 |
0.6 |
| chr17_65527492_65527683 | 0.17 |
CTD-2653B5.1 |
|
6990 |
0.21 |
| chr20_47406387_47406538 | 0.17 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
37958 |
0.18 |
| chr4_16233103_16233254 | 0.17 |
TAPT1-AS1 |
TAPT1 antisense RNA 1 (head to head) |
4510 |
0.23 |
| chr10_67331004_67331235 | 0.17 |
ENSG00000253012 |
. |
138699 |
0.05 |
| chr16_3019207_3020203 | 0.17 |
PAQR4 |
progestin and adipoQ receptor family member IV |
146 |
0.88 |
| chr2_234240529_234240680 | 0.17 |
SAG |
S-antigen; retina and pineal gland (arrestin) |
12410 |
0.13 |
| chr19_8488969_8489220 | 0.17 |
RP11-886P16.6 |
|
3617 |
0.13 |
| chr14_52734332_52734527 | 0.17 |
PTGDR |
prostaglandin D2 receptor (DP) |
2 |
0.99 |
| chr4_126237829_126238914 | 0.17 |
FAT4 |
FAT atypical cadherin 4 |
817 |
0.75 |
| chr19_18451078_18452116 | 0.17 |
PGPEP1 |
pyroglutamyl-peptidase I |
148 |
0.91 |
| chr18_55253153_55254274 | 0.16 |
FECH |
ferrochelatase |
131 |
0.96 |
| chr5_159742923_159743074 | 0.16 |
CCNJL |
cyclin J-like |
3391 |
0.22 |
| chr1_186624970_186625121 | 0.16 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
24514 |
0.25 |
| chr17_76136415_76136566 | 0.16 |
C17orf99 |
chromosome 17 open reading frame 99 |
5944 |
0.12 |
| chr19_862652_862803 | 0.16 |
CFD |
complement factor D (adipsin) |
3037 |
0.11 |
| chr16_85783565_85784591 | 0.16 |
C16orf74 |
chromosome 16 open reading frame 74 |
479 |
0.7 |
| chr11_121315877_121316028 | 0.16 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
6960 |
0.25 |
| chr15_68870208_68871184 | 0.16 |
CORO2B |
coronin, actin binding protein, 2B |
612 |
0.83 |
| chr19_1885517_1885668 | 0.16 |
ABHD17A |
abhydrolase domain containing 17A |
46 |
0.94 |
| chr2_200327762_200327981 | 0.16 |
SATB2 |
SATB homeobox 2 |
1960 |
0.39 |
| chr12_10874089_10875353 | 0.16 |
YBX3 |
Y box binding protein 3 |
1185 |
0.46 |
| chr3_126702505_126703684 | 0.16 |
PLXNA1 |
plexin A1 |
4343 |
0.34 |
| chr1_245316415_245317636 | 0.15 |
KIF26B |
kinesin family member 26B |
1262 |
0.47 |
| chr1_153747814_153748820 | 0.15 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
549 |
0.6 |
| chr1_68298302_68298834 | 0.15 |
GNG12-AS1 |
GNG12 antisense RNA 1 |
576 |
0.46 |
| chr19_48015922_48016172 | 0.15 |
NAPA |
N-ethylmaleimide-sensitive factor attachment protein, alpha |
12 |
0.97 |
| chr11_121327677_121327828 | 0.15 |
RP11-730K11.1 |
|
4030 |
0.27 |
| chr1_85796394_85796580 | 0.15 |
ENSG00000264380 |
. |
46194 |
0.11 |
| chr6_117591445_117591948 | 0.15 |
VGLL2 |
vestigial like 2 (Drosophila) |
4959 |
0.21 |
| chr2_238315058_238315400 | 0.15 |
COL6A3 |
collagen, type VI, alpha 3 |
7562 |
0.22 |
| chr19_9609026_9609526 | 0.15 |
ZNF560 |
zinc finger protein 560 |
7 |
0.98 |
| chr3_3338763_3338914 | 0.15 |
CRBN |
cereblon |
117444 |
0.06 |
| chrX_18372063_18372793 | 0.15 |
SCML2 |
sex comb on midleg-like 2 (Drosophila) |
419 |
0.88 |
| chr17_76164337_76165654 | 0.15 |
SYNGR2 |
synaptogyrin 2 |
244 |
0.87 |
| chr19_50354346_50354967 | 0.14 |
PTOV1 |
prostate tumor overexpressed 1 |
174 |
0.58 |
| chr6_28109319_28110552 | 0.14 |
ZKSCAN8 |
zinc finger with KRAB and SCAN domains 8 |
219 |
0.89 |
| chr20_10530983_10531190 | 0.14 |
JAG1 |
jagged 1 |
112068 |
0.06 |
| chr14_75802545_75802696 | 0.14 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
55724 |
0.1 |
| chr11_48002643_48003787 | 0.14 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
936 |
0.61 |
| chr4_87507373_87507524 | 0.14 |
MAPK10 |
mitogen-activated protein kinase 10 |
7815 |
0.22 |
| chr10_103589679_103590635 | 0.14 |
KCNIP2 |
Kv channel interacting protein 2 |
38 |
0.97 |
| chr15_62684019_62684498 | 0.13 |
RP11-299H22.5 |
|
121379 |
0.05 |
| chr16_30933379_30933820 | 0.13 |
FBXL19 |
F-box and leucine-rich repeat protein 19 |
777 |
0.32 |
| chr5_95214575_95214839 | 0.13 |
ENSG00000250362 |
. |
17217 |
0.13 |
| chr11_62201492_62201643 | 0.13 |
CTD-2531D15.4 |
|
7371 |
0.12 |
| chr22_46067402_46068067 | 0.13 |
ATXN10 |
ataxin 10 |
55 |
0.98 |
| chr1_212809700_212810014 | 0.13 |
RP11-338C15.5 |
|
9744 |
0.14 |
| chr19_50095334_50096202 | 0.13 |
PRR12 |
proline rich 12 |
868 |
0.32 |
| chr5_140625074_140625225 | 0.13 |
PCDHB15 |
protocadherin beta 15 |
2 |
0.94 |
| chr7_28771569_28771914 | 0.13 |
CREB5 |
cAMP responsive element binding protein 5 |
46143 |
0.2 |
| chr16_88278585_88278779 | 0.13 |
ZNF469 |
zinc finger protein 469 |
215197 |
0.02 |
| chr19_1929256_1929607 | 0.13 |
CSNK1G2 |
casein kinase 1, gamma 2 |
11757 |
0.09 |
| chr2_208020804_208020955 | 0.13 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
9416 |
0.21 |
| chr1_1850096_1850795 | 0.13 |
TMEM52 |
transmembrane protein 52 |
209 |
0.9 |
| chr16_29282660_29283032 | 0.13 |
RP11-231C14.6 |
|
40825 |
0.13 |
| chr19_52772900_52773931 | 0.13 |
ZNF766 |
zinc finger protein 766 |
569 |
0.6 |
| chr19_44905150_44906080 | 0.12 |
ZNF285 |
zinc finger protein 285 |
126 |
0.54 |
| chr9_131674102_131674253 | 0.12 |
PHYHD1 |
phytanoyl-CoA dioxygenase domain containing 1 |
8997 |
0.11 |
| chr16_67719587_67719738 | 0.12 |
C16orf86 |
chromosome 16 open reading frame 86 |
18869 |
0.09 |
| chr17_5069290_5069441 | 0.12 |
ZNF594 |
zinc finger protein 594 |
18327 |
0.11 |
| chr11_31892590_31892741 | 0.12 |
PAX6 |
paired box 6 |
53156 |
0.13 |
| chr8_22960171_22961001 | 0.12 |
TNFRSF10C |
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain |
152 |
0.94 |
| chr7_86273005_86273208 | 0.12 |
GRM3 |
glutamate receptor, metabotropic 3 |
124 |
0.98 |
| chr6_42194447_42194755 | 0.12 |
MRPS10 |
mitochondrial ribosomal protein S10 |
8998 |
0.18 |
| chr4_84218783_84218934 | 0.12 |
COQ2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
12791 |
0.23 |
| chr5_140787573_140788174 | 0.12 |
PCDHGB6 |
protocadherin gamma subfamily B, 6 |
103 |
0.89 |
| chr12_3000030_3000938 | 0.12 |
TULP3 |
tubby like protein 3 |
411 |
0.74 |
| chr1_113050561_113051464 | 0.11 |
WNT2B |
wingless-type MMTV integration site family, member 2B |
388 |
0.86 |
| chr6_11432419_11432570 | 0.11 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
49913 |
0.15 |
| chr11_115580481_115580632 | 0.11 |
ENSG00000239153 |
. |
82653 |
0.11 |
| chr16_68280776_68280927 | 0.11 |
PLA2G15 |
phospholipase A2, group XV |
1556 |
0.18 |
| chr3_11651504_11651797 | 0.11 |
VGLL4 |
vestigial like 4 (Drosophila) |
328 |
0.84 |
| chr16_52028189_52028340 | 0.11 |
C16orf97 |
chromosome 16 open reading frame 97 |
33019 |
0.23 |
| chr22_38808730_38808891 | 0.11 |
RP3-434P1.6 |
|
11483 |
0.12 |
| chr19_14448346_14448497 | 0.11 |
CD97 |
CD97 molecule |
43547 |
0.11 |
| chr19_8656644_8657344 | 0.11 |
ADAMTS10 |
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
86 |
0.96 |
| chr1_42230520_42230906 | 0.11 |
ENSG00000264896 |
. |
5901 |
0.3 |
| chr14_35147343_35147800 | 0.11 |
ENSG00000253003 |
. |
2474 |
0.29 |
| chr3_169755608_169756727 | 0.11 |
GPR160 |
G protein-coupled receptor 160 |
241 |
0.91 |
| chr21_15588211_15589196 | 0.11 |
RBM11 |
RNA binding motif protein 11 |
204 |
0.95 |
| chrX_153619023_153619398 | 0.11 |
RPL10 |
ribosomal protein L10 |
6285 |
0.08 |
| chrX_23925220_23926030 | 0.11 |
CXorf58 |
chromosome X open reading frame 58 |
293 |
0.62 |
| chr3_51705298_51706076 | 0.11 |
TEX264 |
testis expressed 264 |
157 |
0.94 |
| chr12_106621577_106621728 | 0.11 |
RP11-651L5.2 |
|
19082 |
0.17 |
| chr9_79520083_79520617 | 0.11 |
PRUNE2 |
prune homolog 2 (Drosophila) |
651 |
0.82 |
| chr14_103394355_103395516 | 0.10 |
AMN |
amnion associated transmembrane protein |
211 |
0.92 |
| chr21_16388752_16388952 | 0.10 |
NRIP1 |
nuclear receptor interacting protein 1 |
14163 |
0.22 |
| chr8_15397591_15398115 | 0.10 |
TUSC3 |
tumor suppressor candidate 3 |
61 |
0.98 |
| chr6_3749707_3750198 | 0.10 |
RP11-420L9.5 |
|
1393 |
0.41 |
| chr1_20799804_20799955 | 0.10 |
CAMK2N1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
12834 |
0.2 |
| chr6_5084934_5086227 | 0.10 |
PPP1R3G |
protein phosphatase 1, regulatory subunit 3G |
140 |
0.97 |
| chr17_45500760_45501815 | 0.10 |
CTD-2026D20.2 |
|
395 |
0.86 |
| chr19_52130500_52130651 | 0.10 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
3013 |
0.15 |
| chr7_93689487_93689638 | 0.10 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
55868 |
0.14 |
| chrX_153961154_153961578 | 0.10 |
GAB3 |
GRB2-associated binding protein 3 |
17966 |
0.12 |
| chr5_124070851_124071458 | 0.10 |
RP11-436H11.5 |
|
286 |
0.82 |
| chr6_167369037_167370142 | 0.10 |
RP11-514O12.4 |
|
23 |
0.9 |
| chr6_149775369_149775520 | 0.10 |
ZC3H12D |
zinc finger CCCH-type containing 12D |
2561 |
0.27 |
| chr10_120788492_120789062 | 0.10 |
NANOS1 |
nanos homolog 1 (Drosophila) |
451 |
0.76 |
| chr19_10341031_10341948 | 0.10 |
ENSG00000264266 |
. |
400 |
0.34 |
| chr19_36360559_36361090 | 0.10 |
APLP1 |
amyloid beta (A4) precursor-like protein 1 |
262 |
0.81 |
| chr11_536799_537414 | 0.10 |
LRRC56 |
leucine rich repeat containing 56 |
421 |
0.62 |
| chr7_139168164_139168924 | 0.10 |
KLRG2 |
killer cell lectin-like receptor subfamily G, member 2 |
86 |
0.97 |
| chr3_55521292_55522040 | 0.09 |
WNT5A-AS1 |
WNT5A antisense RNA 1 |
61 |
0.83 |
| chr3_33397369_33397520 | 0.09 |
ENSG00000252700 |
. |
58198 |
0.12 |
| chrX_153569454_153569812 | 0.09 |
XX-FW83128A1.2 |
|
6246 |
0.1 |
| chr19_13131054_13131205 | 0.09 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
3676 |
0.12 |
| chr16_67197260_67198336 | 0.09 |
HSF4 |
heat shock transcription factor 4 |
510 |
0.52 |
| chr2_239140099_239140700 | 0.09 |
ENSG00000225057 |
. |
74 |
0.5 |
| chr19_50141513_50142109 | 0.09 |
RRAS |
related RAS viral (r-ras) oncogene homolog |
1647 |
0.16 |
| chr9_8857014_8857547 | 0.09 |
PTPRD |
protein tyrosine phosphatase, receptor type, D |
496 |
0.84 |
| chr18_32618955_32619106 | 0.09 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
2294 |
0.44 |
| chr8_17940875_17941847 | 0.09 |
ASAH1 |
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
242 |
0.88 |
| chr15_63793174_63793363 | 0.09 |
USP3 |
ubiquitin specific peptidase 3 |
3525 |
0.29 |
| chr3_36985452_36986747 | 0.09 |
TRANK1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
449 |
0.83 |
| chr8_142245284_142245435 | 0.09 |
SLC45A4 |
solute carrier family 45, member 4 |
4912 |
0.17 |
| chr11_64759386_64759537 | 0.09 |
BATF2 |
basic leucine zipper transcription factor, ATF-like 2 |
1715 |
0.19 |
| chr4_171010363_171010619 | 0.09 |
AADAT |
aminoadipate aminotransferase |
54 |
0.98 |
| chr8_128924040_128924244 | 0.09 |
TMEM75 |
transmembrane protein 75 |
36449 |
0.16 |
| chr4_26263054_26263205 | 0.09 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
11100 |
0.31 |
| chr6_41410877_41411028 | 0.09 |
ENSG00000238867 |
. |
87765 |
0.06 |
| chr9_130966078_130966935 | 0.09 |
CIZ1 |
CDKN1A interacting zinc finger protein 1 |
156 |
0.85 |
| chr6_158067098_158067249 | 0.09 |
ZDHHC14 |
zinc finger, DHHC-type containing 14 |
53132 |
0.16 |
| chr2_231733015_231733166 | 0.09 |
ITM2C |
integral membrane protein 2C |
2711 |
0.25 |
| chr16_66558097_66558248 | 0.09 |
TK2 |
thymidine kinase 2, mitochondrial |
10486 |
0.1 |
| chr1_226822390_226822541 | 0.09 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
24906 |
0.19 |
| chr15_37391316_37391907 | 0.09 |
MEIS2 |
Meis homeobox 2 |
14 |
0.54 |
| chr1_33547337_33547515 | 0.09 |
ADC |
arginine decarboxylase |
425 |
0.84 |
| chr6_168078748_168078899 | 0.09 |
AL009178.1 |
Uncharacterized protein; cDNA FLJ43200 fis, clone FEBRA2007793 |
117387 |
0.06 |
| chr17_7608077_7608845 | 0.09 |
EFNB3 |
ephrin-B3 |
59 |
0.94 |
| chr1_155011778_155011941 | 0.09 |
DCST1 |
DC-STAMP domain containing 1 |
5413 |
0.08 |
| chr20_44650596_44650981 | 0.09 |
RP11-465L10.10 |
|
76 |
0.85 |
| chr8_102020336_102020487 | 0.09 |
ENSG00000252736 |
. |
45140 |
0.13 |
| chr12_133137038_133137885 | 0.09 |
P2RX2 |
purinergic receptor P2X, ligand-gated ion channel, 2 |
57905 |
0.1 |
| chr12_95597108_95597270 | 0.09 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
13966 |
0.18 |
| chrX_153639895_153640843 | 0.08 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
51 |
0.66 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.2 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.1 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.4 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.0 | 0.1 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.0 | 0.0 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0090382 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.0 | GO:0032048 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.2 | GO:0030818 | negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 0.3 | GO:0042133 | neurotransmitter metabolic process(GO:0042133) |
| 0.0 | 0.0 | GO:0021508 | floor plate formation(GO:0021508) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.2 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |