Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
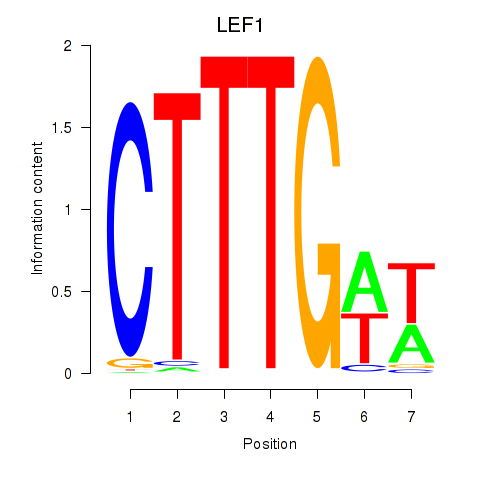
Results for LEF1
Z-value: 0.98
Transcription factors associated with LEF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LEF1
|
ENSG00000138795.5 | lymphoid enhancer binding factor 1 |
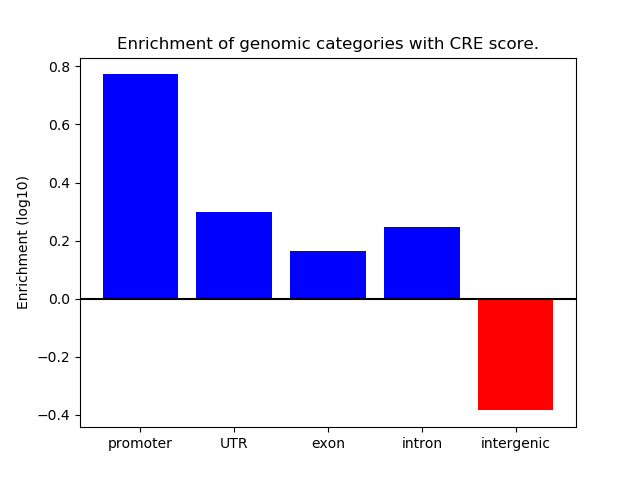
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_109055624_109055775 | LEF1 | 31758 | 0.182969 | -0.45 | 2.2e-01 | Click! |
| chr4_109062164_109062315 | LEF1 | 25218 | 0.197824 | 0.40 | 2.8e-01 | Click! |
| chr4_109076513_109076664 | LEF1 | 10869 | 0.226088 | 0.40 | 2.9e-01 | Click! |
| chr4_109012857_109013008 | LEF1 | 74525 | 0.091399 | 0.38 | 3.2e-01 | Click! |
| chr4_109037667_109037818 | LEF1 | 49715 | 0.140440 | -0.33 | 3.9e-01 | Click! |
Activity of the LEF1 motif across conditions
Conditions sorted by the z-value of the LEF1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
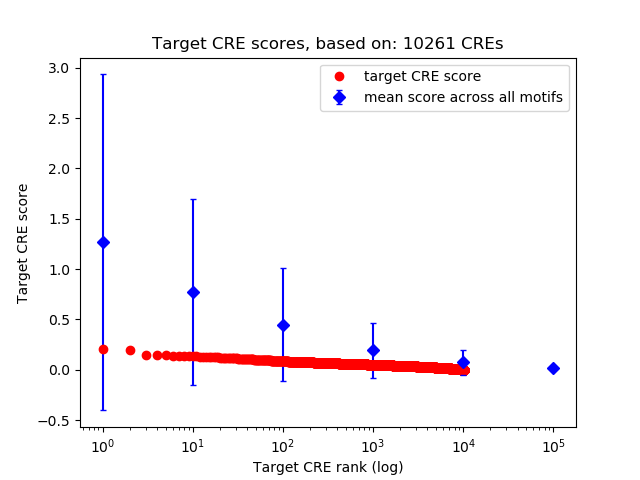
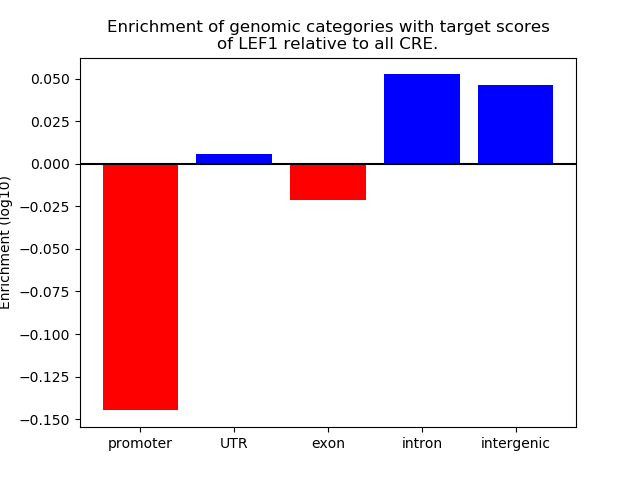
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_169652381_169652976 | 0.20 |
THBS2 |
thrombospondin 2 |
432 |
0.89 |
| chr14_23447268_23447766 | 0.20 |
AJUBA |
ajuba LIM protein |
571 |
0.5 |
| chr15_30112674_30113382 | 0.15 |
TJP1 |
tight junction protein 1 |
723 |
0.64 |
| chr10_122451080_122451231 | 0.15 |
WDR11-AS1 |
WDR11 antisense RNA 1 |
85241 |
0.09 |
| chr5_32985210_32985395 | 0.14 |
AC026703.1 |
|
196357 |
0.03 |
| chr13_60593844_60594053 | 0.14 |
DIAPH3-AS1 |
DIAPH3 antisense RNA 1 |
6508 |
0.25 |
| chr1_7439604_7439992 | 0.14 |
RP3-453P22.2 |
|
9681 |
0.21 |
| chr5_175460576_175460928 | 0.14 |
THOC3 |
THO complex 3 |
931 |
0.5 |
| chr14_51909906_51910057 | 0.14 |
FRMD6 |
FERM domain containing 6 |
45874 |
0.16 |
| chr2_189158808_189158959 | 0.13 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
338 |
0.89 |
| chr16_88135173_88135324 | 0.13 |
BANP |
BTG3 associated nuclear protein |
131624 |
0.05 |
| chr3_123699067_123699263 | 0.13 |
ROPN1 |
rhophilin associated tail protein 1 |
11034 |
0.2 |
| chr4_86700210_86700446 | 0.13 |
ARHGAP24 |
Rho GTPase activating protein 24 |
469 |
0.88 |
| chr2_242802004_242802249 | 0.13 |
PDCD1 |
programmed cell death 1 |
1066 |
0.37 |
| chr3_192634406_192635568 | 0.13 |
MB21D2 |
Mab-21 domain containing 2 |
963 |
0.71 |
| chr10_94725160_94725374 | 0.13 |
EXOC6 |
exocyst complex component 6 |
31964 |
0.2 |
| chr21_32533012_32533247 | 0.12 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
30590 |
0.23 |
| chr18_67714217_67714443 | 0.12 |
RTTN |
rotatin |
26376 |
0.25 |
| chr4_52904501_52904984 | 0.12 |
SGCB |
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
94 |
0.97 |
| chr10_105614245_105615135 | 0.12 |
SH3PXD2A |
SH3 and PX domains 2A |
474 |
0.82 |
| chr2_24398914_24399065 | 0.12 |
FAM228A |
family with sequence similarity 228, member A |
635 |
0.51 |
| chr4_20260773_20260924 | 0.12 |
SLIT2 |
slit homolog 2 (Drosophila) |
4305 |
0.36 |
| chr8_26165121_26165272 | 0.12 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
14197 |
0.25 |
| chr11_26211058_26211209 | 0.12 |
ANO3 |
anoctamin 3 |
304 |
0.95 |
| chr7_14417428_14417579 | 0.12 |
ENSG00000252374 |
. |
367115 |
0.01 |
| chr7_96653221_96653741 | 0.11 |
DLX5 |
distal-less homeobox 5 |
781 |
0.6 |
| chr9_113530116_113530340 | 0.11 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
7399 |
0.3 |
| chr4_140807822_140807973 | 0.11 |
MAML3 |
mastermind-like 3 (Drosophila) |
3309 |
0.35 |
| chr11_35642492_35642706 | 0.11 |
FJX1 |
four jointed box 1 (Drosophila) |
2864 |
0.32 |
| chr15_40630709_40631123 | 0.11 |
C15orf52 |
chromosome 15 open reading frame 52 |
582 |
0.52 |
| chrX_66767815_66768085 | 0.11 |
AR |
androgen receptor |
2964 |
0.43 |
| chr10_32202333_32202484 | 0.11 |
ARHGAP12 |
Rho GTPase activating protein 12 |
4606 |
0.3 |
| chr15_92080010_92080161 | 0.11 |
ENSG00000238981 |
. |
298524 |
0.01 |
| chr15_96212773_96212983 | 0.11 |
ENSG00000222076 |
. |
76155 |
0.13 |
| chr5_52489907_52490058 | 0.11 |
MOCS2 |
molybdenum cofactor synthesis 2 |
84089 |
0.09 |
| chr2_238318512_238318719 | 0.11 |
COL6A3 |
collagen, type VI, alpha 3 |
4176 |
0.24 |
| chr2_220942198_220942388 | 0.11 |
ENSG00000221199 |
. |
114306 |
0.06 |
| chr11_64333824_64334194 | 0.11 |
SLC22A11 |
solute carrier family 22 (organic anion/urate transporter), member 11 |
1063 |
0.49 |
| chr6_81938853_81939008 | 0.11 |
RP1-300G12.2 |
|
305700 |
0.01 |
| chr2_11887384_11888228 | 0.11 |
LPIN1 |
lipin 1 |
1042 |
0.41 |
| chr4_54581434_54581585 | 0.11 |
LNX1 |
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
13937 |
0.22 |
| chr2_12859088_12859514 | 0.10 |
TRIB2 |
tribbles pseudokinase 2 |
937 |
0.67 |
| chr3_98666159_98666310 | 0.10 |
ENSG00000207331 |
. |
38931 |
0.15 |
| chrX_34673509_34673660 | 0.10 |
TMEM47 |
transmembrane protein 47 |
1821 |
0.53 |
| chr3_112961154_112961680 | 0.10 |
BOC |
BOC cell adhesion associated, oncogene regulated |
26219 |
0.19 |
| chr12_13359480_13359859 | 0.10 |
EMP1 |
epithelial membrane protein 1 |
4776 |
0.29 |
| chr2_238321225_238321570 | 0.10 |
COL6A3 |
collagen, type VI, alpha 3 |
1394 |
0.45 |
| chr4_158916155_158916306 | 0.10 |
FAM198B |
family with sequence similarity 198, member B |
164603 |
0.03 |
| chr3_149356356_149356942 | 0.10 |
WWTR1-IT1 |
WWTR1 intronic transcript 1 (non-protein coding) |
11322 |
0.17 |
| chr16_73236593_73236744 | 0.10 |
C16orf47 |
chromosome 16 open reading frame 47 |
58322 |
0.16 |
| chr2_58250808_58250969 | 0.10 |
VRK2 |
vaccinia related kinase 2 |
22841 |
0.23 |
| chr9_4490575_4491197 | 0.10 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
442 |
0.88 |
| chr2_15310036_15310187 | 0.10 |
NBAS |
neuroblastoma amplified sequence |
20410 |
0.29 |
| chr18_49683791_49683942 | 0.10 |
DCC |
deleted in colorectal carcinoma |
182676 |
0.03 |
| chr15_96869332_96869483 | 0.10 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
240 |
0.83 |
| chr2_119603311_119603908 | 0.10 |
EN1 |
engrailed homeobox 1 |
1645 |
0.51 |
| chr10_11371205_11371471 | 0.10 |
CELF2-AS1 |
CELF2 antisense RNA 1 |
9491 |
0.29 |
| chr12_110723019_110723319 | 0.10 |
ATP2A2 |
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
3683 |
0.28 |
| chr3_33686746_33686949 | 0.10 |
CLASP2 |
cytoplasmic linker associated protein 2 |
40 |
0.99 |
| chrX_54947220_54948504 | 0.10 |
TRO |
trophinin |
41 |
0.97 |
| chr15_97433403_97433676 | 0.10 |
ENSG00000223120 |
. |
48240 |
0.17 |
| chr18_53089837_53090372 | 0.10 |
TCF4 |
transcription factor 4 |
361 |
0.89 |
| chr12_88972313_88972545 | 0.10 |
KITLG |
KIT ligand |
1809 |
0.39 |
| chr7_137567705_137567869 | 0.09 |
DGKI |
diacylglycerol kinase, iota |
35949 |
0.17 |
| chr10_124221611_124222138 | 0.09 |
HTRA1 |
HtrA serine peptidase 1 |
833 |
0.61 |
| chr15_89181571_89183025 | 0.09 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
114 |
0.96 |
| chr6_100316163_100316348 | 0.09 |
MCHR2-AS1 |
MCHR2 antisense RNA 1 |
125565 |
0.05 |
| chr5_66611355_66611669 | 0.09 |
CD180 |
CD180 molecule |
118885 |
0.06 |
| chr5_173846066_173846362 | 0.09 |
CTC-229L21.1 |
|
246110 |
0.02 |
| chr1_197163991_197164285 | 0.09 |
ZBTB41 |
zinc finger and BTB domain containing 41 |
5534 |
0.22 |
| chr2_36796670_36797008 | 0.09 |
FEZ2 |
fasciculation and elongation protein zeta 2 (zygin II) |
11631 |
0.19 |
| chr7_113722711_113723554 | 0.09 |
FOXP2 |
forkhead box P2 |
3483 |
0.3 |
| chr13_24144563_24144801 | 0.09 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
121 |
0.98 |
| chr6_111949604_111949832 | 0.09 |
ENSG00000239015 |
. |
17912 |
0.16 |
| chr22_23863444_23864162 | 0.09 |
AP000345.2 |
|
45448 |
0.12 |
| chr20_43976437_43976958 | 0.09 |
SDC4 |
syndecan 4 |
367 |
0.79 |
| chr17_42004293_42004804 | 0.09 |
RP11-527L4.5 |
|
4179 |
0.12 |
| chr6_143994597_143994872 | 0.09 |
PHACTR2 |
phosphatase and actin regulator 2 |
4368 |
0.31 |
| chr7_841605_841901 | 0.09 |
SUN1 |
Sad1 and UNC84 domain containing 1 |
13775 |
0.17 |
| chr3_125690962_125691349 | 0.09 |
ROPN1B |
rhophilin associated tail protein 1B |
3098 |
0.23 |
| chr15_60684685_60685107 | 0.09 |
ANXA2 |
annexin A2 |
1400 |
0.54 |
| chr10_54075051_54075395 | 0.09 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
1167 |
0.43 |
| chr4_103737426_103737626 | 0.09 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
9173 |
0.15 |
| chr10_25137198_25137349 | 0.09 |
ENSG00000240294 |
. |
60468 |
0.13 |
| chr7_27218725_27218915 | 0.09 |
RP1-170O19.20 |
Uncharacterized protein |
812 |
0.27 |
| chr8_122360745_122360896 | 0.09 |
ENSG00000221644 |
. |
161688 |
0.04 |
| chr3_159506026_159506232 | 0.09 |
IQCJ-SCHIP1-AS1 |
IQCJ-SCHIP1 readthrough antisense RNA 1 |
19728 |
0.15 |
| chr17_76785896_76786047 | 0.09 |
CYTH1 |
cytohesin 1 |
7592 |
0.18 |
| chr4_101342464_101342615 | 0.09 |
EMCN |
endomucin |
53695 |
0.17 |
| chr20_51475685_51475852 | 0.09 |
TSHZ2 |
teashirt zinc finger homeobox 2 |
113178 |
0.07 |
| chr16_89990293_89990742 | 0.09 |
TUBB3 |
Tubulin beta-3 chain |
743 |
0.44 |
| chr11_46670801_46671004 | 0.09 |
ATG13 |
autophagy related 13 |
10070 |
0.15 |
| chr7_22536430_22536632 | 0.09 |
STEAP1B |
STEAP family member 1B |
3265 |
0.31 |
| chr10_54079102_54079253 | 0.09 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
5121 |
0.26 |
| chr11_12836551_12836790 | 0.09 |
RP11-47J17.3 |
|
8544 |
0.22 |
| chr10_119564055_119564206 | 0.09 |
RP11-354M20.3 |
|
212868 |
0.02 |
| chr7_134464463_134465343 | 0.09 |
CALD1 |
caldesmon 1 |
474 |
0.89 |
| chr4_114898942_114899097 | 0.09 |
ARSJ |
arylsulfatase family, member J |
1133 |
0.61 |
| chr1_214397865_214398571 | 0.09 |
SMYD2 |
SET and MYND domain containing 2 |
56358 |
0.16 |
| chr1_156087932_156088663 | 0.09 |
LMNA |
lamin A/C |
3784 |
0.13 |
| chr5_169398695_169398846 | 0.09 |
FAM196B |
family with sequence similarity 196, member B |
8974 |
0.26 |
| chr22_45607681_45607964 | 0.09 |
KIAA0930 |
KIAA0930 |
476 |
0.78 |
| chr18_7579845_7579996 | 0.09 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
12103 |
0.29 |
| chr7_115116914_115117103 | 0.09 |
ENSG00000202377 |
. |
104488 |
0.08 |
| chr2_151260036_151260226 | 0.09 |
RND3 |
Rho family GTPase 3 |
81765 |
0.12 |
| chr7_100464814_100466140 | 0.09 |
TRIP6 |
thyroid hormone receptor interactor 6 |
717 |
0.47 |
| chr13_110958781_110959436 | 0.08 |
COL4A1 |
collagen, type IV, alpha 1 |
370 |
0.6 |
| chr9_111206334_111206588 | 0.08 |
ENSG00000222512 |
. |
85252 |
0.11 |
| chr3_149371362_149371801 | 0.08 |
WWTR1-AS1 |
WWTR1 antisense RNA 1 |
3226 |
0.2 |
| chr11_130319616_130319897 | 0.08 |
ADAMTS15 |
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
887 |
0.67 |
| chr8_142215547_142216678 | 0.08 |
DENND3 |
DENN/MADD domain containing 3 |
13682 |
0.16 |
| chr3_49237110_49237583 | 0.08 |
CCDC36 |
coiled-coil domain containing 36 |
403 |
0.74 |
| chr1_68280586_68280737 | 0.08 |
GNG12-AS1 |
GNG12 antisense RNA 1 |
17325 |
0.19 |
| chr7_80546807_80546971 | 0.08 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
1610 |
0.56 |
| chr4_27086932_27087083 | 0.08 |
STIM2 |
stromal interaction molecule 2 |
76912 |
0.12 |
| chr8_32184476_32184627 | 0.08 |
ENSG00000200246 |
. |
70539 |
0.11 |
| chr4_79141777_79141928 | 0.08 |
FRAS1 |
Fraser syndrome 1 |
24533 |
0.22 |
| chr10_33619964_33620115 | 0.08 |
NRP1 |
neuropilin 1 |
3271 |
0.33 |
| chr2_237652692_237652863 | 0.08 |
ACKR3 |
atypical chemokine receptor 3 |
174493 |
0.03 |
| chr10_88518871_88519024 | 0.08 |
BMPR1A |
bone morphogenetic protein receptor, type IA |
2540 |
0.23 |
| chr5_167899041_167899382 | 0.08 |
RARS |
arginyl-tRNA synthetase |
14239 |
0.2 |
| chr17_78699090_78699345 | 0.08 |
RP11-28G8.1 |
|
80215 |
0.09 |
| chr2_20348576_20349061 | 0.08 |
ENSG00000266059 |
. |
26289 |
0.15 |
| chr17_67056652_67056803 | 0.08 |
ABCA9 |
ATP-binding cassette, sub-family A (ABC1), member 9 |
320 |
0.9 |
| chr17_77223432_77223590 | 0.08 |
RBFOX3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
8376 |
0.27 |
| chr13_75018049_75018424 | 0.08 |
LINC00381 |
long intergenic non-protein coding RNA 381 |
24926 |
0.22 |
| chrX_46186736_46187042 | 0.08 |
KRBOX4 |
KRAB box domain containing 4 |
119403 |
0.06 |
| chr3_51377510_51377669 | 0.08 |
MANF |
mesencephalic astrocyte-derived neurotrophic factor |
44889 |
0.15 |
| chr12_57522507_57523183 | 0.08 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
38 |
0.64 |
| chr7_38546645_38546796 | 0.08 |
ENSG00000241756 |
. |
24219 |
0.23 |
| chr3_98603544_98603695 | 0.08 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
16396 |
0.17 |
| chr13_79182716_79183828 | 0.08 |
POU4F1 |
POU class 4 homeobox 1 |
5599 |
0.19 |
| chr15_37180001_37180243 | 0.08 |
ENSG00000212511 |
. |
35281 |
0.22 |
| chr14_37131505_37132074 | 0.08 |
PAX9 |
paired box 9 |
10 |
0.98 |
| chr20_61656853_61657255 | 0.08 |
BHLHE23 |
basic helix-loop-helix family, member e23 |
18667 |
0.17 |
| chr15_61012825_61012976 | 0.08 |
ENSG00000212625 |
. |
16068 |
0.2 |
| chr9_36029255_36029406 | 0.08 |
RECK |
reversion-inducing-cysteine-rich protein with kazal motifs |
7100 |
0.21 |
| chr3_191047655_191048448 | 0.08 |
UTS2B |
urotensin 2B |
211 |
0.91 |
| chr18_33878744_33878957 | 0.08 |
FHOD3 |
formin homology 2 domain containing 3 |
1051 |
0.65 |
| chr20_30457693_30458808 | 0.08 |
DUSP15 |
dual specificity phosphatase 15 |
125 |
0.67 |
| chr3_149686814_149687069 | 0.08 |
PFN2 |
profilin 2 |
1247 |
0.4 |
| chr3_112358958_112359194 | 0.08 |
CCDC80 |
coiled-coil domain containing 80 |
1040 |
0.62 |
| chr3_156794251_156794402 | 0.08 |
ENSG00000222499 |
. |
59195 |
0.11 |
| chr12_80663224_80663505 | 0.08 |
OTOGL |
otogelin-like |
60131 |
0.13 |
| chr5_35797219_35797401 | 0.08 |
SPEF2 |
sperm flagellar 2 |
18040 |
0.19 |
| chr5_158203015_158203166 | 0.08 |
CTD-2363C16.1 |
|
206924 |
0.02 |
| chr12_47358106_47358377 | 0.08 |
PCED1B |
PC-esterase domain containing 1B |
115145 |
0.06 |
| chr2_38828054_38829790 | 0.08 |
AC011247.3 |
|
182 |
0.89 |
| chr11_65158052_65158759 | 0.08 |
FRMD8 |
FERM domain containing 8 |
4006 |
0.13 |
| chr6_140323383_140323534 | 0.08 |
ENSG00000252107 |
. |
156373 |
0.04 |
| chr8_17611273_17611424 | 0.08 |
MTUS1 |
microtubule associated tumor suppressor 1 |
2142 |
0.28 |
| chr12_12972772_12973017 | 0.08 |
DDX47 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
6600 |
0.18 |
| chrX_45661389_45661688 | 0.08 |
ENSG00000207725 |
. |
55008 |
0.14 |
| chr1_216705124_216705304 | 0.08 |
USH2A |
Usher syndrome 2A (autosomal recessive, mild) |
108476 |
0.08 |
| chr5_9544374_9544669 | 0.08 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
1666 |
0.36 |
| chr5_102096978_102097129 | 0.08 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
6006 |
0.34 |
| chr9_21576816_21576971 | 0.08 |
MIR31HG |
MIR31 host gene (non-protein coding) |
17225 |
0.18 |
| chr6_154650904_154651062 | 0.08 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
232 |
0.96 |
| chr6_6331737_6331888 | 0.08 |
F13A1 |
coagulation factor XIII, A1 polypeptide |
10566 |
0.3 |
| chr1_195022621_195022810 | 0.08 |
ENSG00000265108 |
. |
73096 |
0.14 |
| chr2_182819535_182819810 | 0.08 |
PPP1R1C |
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
30879 |
0.17 |
| chr4_138451566_138451717 | 0.08 |
PCDH18 |
protocadherin 18 |
1924 |
0.52 |
| chr4_24472696_24473642 | 0.08 |
ENSG00000265001 |
. |
41714 |
0.15 |
| chr17_57704803_57704954 | 0.08 |
CLTC |
clathrin, heavy chain (Hc) |
7585 |
0.21 |
| chr5_174178214_174178699 | 0.08 |
ENSG00000266890 |
. |
281 |
0.94 |
| chr2_187214072_187214223 | 0.08 |
ZC3H15 |
zinc finger CCCH-type containing 15 |
136736 |
0.05 |
| chr12_6310274_6310804 | 0.08 |
CD9 |
CD9 molecule |
576 |
0.75 |
| chr4_128424253_128424404 | 0.08 |
INTU |
inturned planar cell polarity protein |
120098 |
0.06 |
| chr6_167171492_167171681 | 0.08 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
187 |
0.97 |
| chr9_134282757_134283214 | 0.08 |
PRRC2B |
proline-rich coiled-coil 2B |
13505 |
0.2 |
| chr15_57946603_57946754 | 0.08 |
MYZAP |
myocardial zonula adherens protein |
20816 |
0.22 |
| chr5_53753293_53753597 | 0.08 |
HSPB3 |
heat shock 27kDa protein 3 |
2000 |
0.44 |
| chr10_79647530_79647963 | 0.08 |
AL391421.1 |
Uncharacterized protein; cDNA FLJ43696 fis, clone TBAES2007964 |
21113 |
0.15 |
| chr10_14051355_14052213 | 0.08 |
FRMD4A |
FERM domain containing 4A |
1252 |
0.51 |
| chr10_13931045_13931263 | 0.08 |
FRMD4A |
FERM domain containing 4A |
30242 |
0.2 |
| chr2_74606600_74607395 | 0.08 |
DCTN1 |
dynactin 1 |
413 |
0.73 |
| chr18_32291325_32291476 | 0.08 |
DTNA |
dystrobrevin, alpha |
1139 |
0.62 |
| chr6_106207677_106207828 | 0.08 |
ENSG00000200198 |
. |
144496 |
0.05 |
| chr13_21287159_21287369 | 0.08 |
ENSG00000265710 |
. |
9422 |
0.2 |
| chr15_67021750_67022170 | 0.08 |
SMAD6 |
SMAD family member 6 |
17925 |
0.24 |
| chr1_216897072_216897310 | 0.08 |
ESRRG |
estrogen-related receptor gamma |
396 |
0.93 |
| chr17_59882538_59882689 | 0.08 |
BRIP1 |
BRCA1 interacting protein C-terminal helicase 1 |
28772 |
0.2 |
| chr12_57527559_57527864 | 0.08 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
1789 |
0.2 |
| chr4_30724120_30724271 | 0.08 |
PCDH7 |
protocadherin 7 |
218 |
0.97 |
| chr9_112091327_112091518 | 0.08 |
ENSG00000207329 |
. |
4457 |
0.22 |
| chr18_56528219_56528370 | 0.08 |
ZNF532 |
zinc finger protein 532 |
1538 |
0.39 |
| chr3_29377379_29377667 | 0.08 |
ENSG00000216169 |
. |
33389 |
0.18 |
| chr12_114841509_114841660 | 0.08 |
TBX5 |
T-box 5 |
119 |
0.97 |
| chr21_28214648_28215240 | 0.08 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
393 |
0.9 |
| chr17_78166034_78166254 | 0.08 |
CARD14 |
caspase recruitment domain family, member 14 |
8170 |
0.14 |
| chr16_67282110_67283381 | 0.07 |
SLC9A5 |
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
108 |
0.89 |
| chr10_52835110_52835261 | 0.07 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
1251 |
0.52 |
| chr22_46479348_46479691 | 0.07 |
FLJ27365 |
hsa-mir-4763 |
2364 |
0.16 |
| chr7_55089056_55089207 | 0.07 |
EGFR |
epidermal growth factor receptor |
2320 |
0.44 |
| chr15_34728474_34728854 | 0.07 |
GOLGA8A |
golgin A8 family, member A |
28776 |
0.13 |
| chr9_18476160_18476529 | 0.07 |
ADAMTSL1 |
ADAMTS-like 1 |
2113 |
0.45 |
| chr16_29151612_29151763 | 0.07 |
CTB-134H23.3 |
|
32921 |
0.13 |
| chr9_14348492_14348672 | 0.07 |
RP11-120J1.1 |
|
1263 |
0.57 |
| chr1_178096168_178096398 | 0.07 |
RASAL2 |
RAS protein activator like 2 |
33007 |
0.24 |
| chr10_14050350_14050950 | 0.07 |
FRMD4A |
FERM domain containing 4A |
118 |
0.97 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 0.2 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.2 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0035844 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0003207 | cardiac chamber formation(GO:0003207) cardiac ventricle formation(GO:0003211) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0050942 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.0 | GO:0043301 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.0 | 0.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.0 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:1904238 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0006533 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.2 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.0 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.0 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.0 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.0 | GO:0044217 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.3 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.0 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.0 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.0 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |