Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
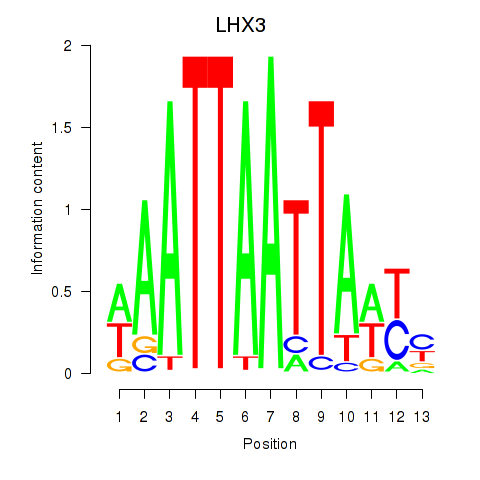
Results for LHX3
Z-value: 1.23
Transcription factors associated with LHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX3
|
ENSG00000107187.11 | LIM homeobox 3 |
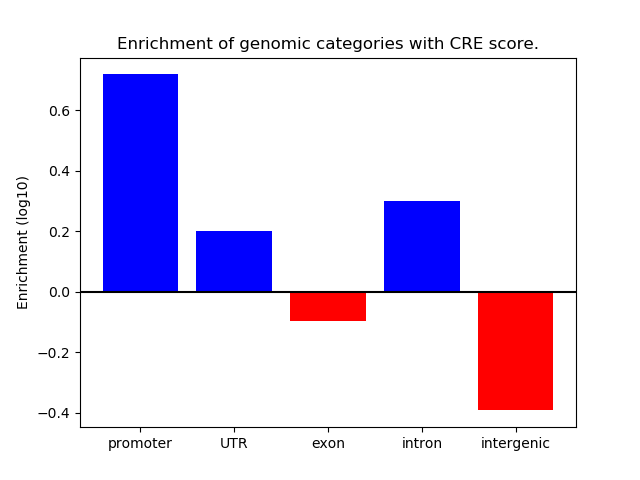
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_139091429_139091580 | LHX3 | 3500 | 0.229468 | 0.15 | 6.9e-01 | Click! |
| chr9_139090516_139090790 | LHX3 | 4351 | 0.212434 | -0.00 | 1.0e+00 | Click! |
Activity of the LHX3 motif across conditions
Conditions sorted by the z-value of the LHX3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
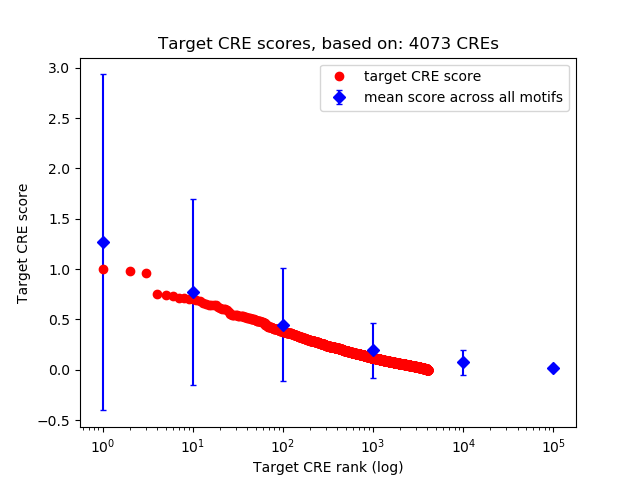
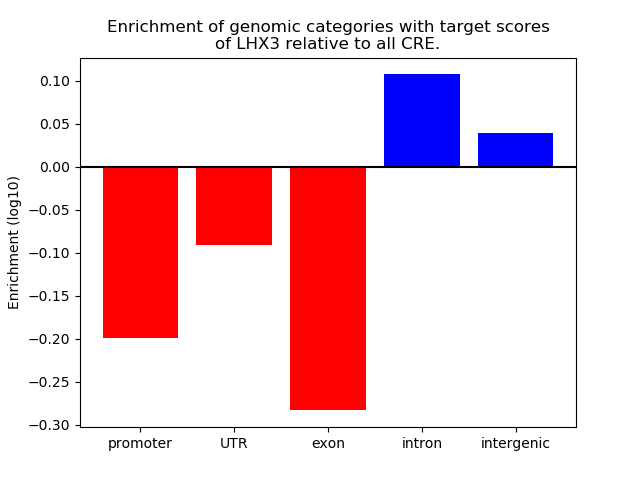
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_41215270_41215769 | 1.01 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
956 |
0.59 |
| chr8_116675050_116675578 | 0.98 |
TRPS1 |
trichorhinophalangeal syndrome I |
1409 |
0.6 |
| chr9_89952337_89952906 | 0.96 |
ENSG00000212421 |
. |
77256 |
0.11 |
| chr12_66286178_66286489 | 0.75 |
RP11-366L20.2 |
Uncharacterized protein |
10386 |
0.18 |
| chr11_122311553_122311819 | 0.74 |
ENSG00000252776 |
. |
8232 |
0.23 |
| chr17_35851874_35852313 | 0.73 |
DUSP14 |
dual specificity phosphatase 14 |
523 |
0.79 |
| chr3_79066759_79066910 | 0.72 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
860 |
0.75 |
| chr6_27060216_27060434 | 0.71 |
ENSG00000222800 |
. |
17434 |
0.13 |
| chrX_114829080_114829277 | 0.70 |
PLS3 |
plastin 3 |
1313 |
0.48 |
| chr7_98970111_98970529 | 0.70 |
ARPC1B |
actin related protein 2/3 complex, subunit 1B, 41kDa |
1552 |
0.28 |
| chr18_42313149_42313495 | 0.70 |
SETBP1 |
SET binding protein 1 |
36193 |
0.24 |
| chr6_147234783_147235185 | 0.69 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
2037 |
0.48 |
| chr3_33318777_33319866 | 0.67 |
FBXL2 |
F-box and leucine-rich repeat protein 2 |
353 |
0.91 |
| chr20_46412966_46413363 | 0.66 |
SULF2 |
sulfatase 2 |
1069 |
0.59 |
| chr4_108746841_108747185 | 0.65 |
SGMS2 |
sphingomyelin synthase 2 |
649 |
0.76 |
| chr4_140808579_140808865 | 0.64 |
MAML3 |
mastermind-like 3 (Drosophila) |
2484 |
0.4 |
| chr10_33798417_33798757 | 0.64 |
NRP1 |
neuropilin 1 |
173397 |
0.03 |
| chr9_97812256_97812766 | 0.64 |
ENSG00000207563 |
. |
34979 |
0.12 |
| chr13_80912572_80913077 | 0.62 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
970 |
0.67 |
| chr3_60921693_60921844 | 0.62 |
ENSG00000212211 |
. |
79491 |
0.12 |
| chr12_104443157_104443419 | 0.60 |
GLT8D2 |
glycosyltransferase 8 domain containing 2 |
626 |
0.69 |
| chr3_120168709_120169860 | 0.60 |
FSTL1 |
follistatin-like 1 |
554 |
0.84 |
| chrX_139792263_139792666 | 0.60 |
LINC00632 |
long intergenic non-protein coding RNA 632 |
532 |
0.82 |
| chr19_52452059_52452669 | 0.59 |
HCCAT3 |
hepatocellular carcinoma associated transcript 3 (non-protein coding) |
23 |
0.96 |
| chr1_120191467_120191618 | 0.57 |
ZNF697 |
zinc finger protein 697 |
1146 |
0.52 |
| chr12_109116833_109117152 | 0.55 |
CORO1C |
coronin, actin binding protein, 1C |
7335 |
0.16 |
| chr5_120114861_120115414 | 0.55 |
ENSG00000222609 |
. |
68602 |
0.14 |
| chr8_108506884_108507225 | 0.54 |
ANGPT1 |
angiopoietin 1 |
169 |
0.98 |
| chr3_12235413_12235694 | 0.54 |
TIMP4 |
TIMP metallopeptidase inhibitor 4 |
34702 |
0.2 |
| chrX_39548256_39548819 | 0.54 |
ENSG00000263730 |
. |
28067 |
0.24 |
| chr5_124748322_124748473 | 0.54 |
ENSG00000222107 |
. |
61573 |
0.16 |
| chr13_24146066_24146217 | 0.54 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
1338 |
0.59 |
| chr4_25238162_25238313 | 0.53 |
PI4K2B |
phosphatidylinositol 4-kinase type 2 beta |
2640 |
0.35 |
| chr11_111849006_111849202 | 0.53 |
DIXDC1 |
DIX domain containing 1 |
1071 |
0.39 |
| chr8_107738861_107739281 | 0.53 |
OXR1 |
oxidation resistance 1 |
658 |
0.71 |
| chr5_1883178_1883404 | 0.53 |
IRX4 |
iroquois homeobox 4 |
411 |
0.66 |
| chr18_53089837_53090372 | 0.53 |
TCF4 |
transcription factor 4 |
361 |
0.89 |
| chr1_245361051_245361416 | 0.53 |
RP11-62I21.1 |
|
36570 |
0.15 |
| chr13_79182716_79183828 | 0.52 |
POU4F1 |
POU class 4 homeobox 1 |
5599 |
0.19 |
| chr3_100785098_100785499 | 0.52 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
72939 |
0.11 |
| chr11_89113858_89114276 | 0.51 |
NOX4 |
NADPH oxidase 4 |
109833 |
0.07 |
| chr6_28442401_28443147 | 0.51 |
ZSCAN23 |
zinc finger and SCAN domain containing 23 |
31530 |
0.13 |
| chr14_52821573_52821780 | 0.51 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
40563 |
0.18 |
| chr7_28726429_28726958 | 0.51 |
CREB5 |
cAMP responsive element binding protein 5 |
1095 |
0.68 |
| chr1_64197696_64197847 | 0.51 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
41922 |
0.17 |
| chr14_75039149_75039311 | 0.51 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
39545 |
0.13 |
| chr22_18575359_18575510 | 0.51 |
XXbac-B476C20.9 |
|
13784 |
0.14 |
| chr4_54424539_54424751 | 0.51 |
LNX1 |
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
261 |
0.93 |
| chr13_45242490_45242715 | 0.49 |
ENSG00000238932 |
. |
39793 |
0.2 |
| chr10_33621519_33621932 | 0.49 |
NRP1 |
neuropilin 1 |
1585 |
0.49 |
| chr9_93557702_93558119 | 0.49 |
SYK |
spleen tyrosine kinase |
6159 |
0.34 |
| chr15_99444137_99444307 | 0.49 |
RP11-654A16.1 |
|
7458 |
0.22 |
| chr2_164590425_164590732 | 0.49 |
FIGN |
fidgetin |
1939 |
0.52 |
| chr5_72327202_72327380 | 0.48 |
ENSG00000200833 |
. |
68609 |
0.09 |
| chr7_110879115_110879283 | 0.48 |
ENSG00000238922 |
. |
129786 |
0.05 |
| chr16_53244996_53245358 | 0.48 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
2814 |
0.29 |
| chr14_75038296_75038530 | 0.48 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
40362 |
0.13 |
| chr15_45940670_45940821 | 0.48 |
SQRDL |
sulfide quinone reductase-like (yeast) |
2666 |
0.28 |
| chr1_182365770_182365966 | 0.48 |
TEDDM1 |
transmembrane epididymal protein 1 |
3883 |
0.23 |
| chr2_38295840_38295991 | 0.47 |
RMDN2-AS1 |
RMDN2 antisense RNA 1 |
1731 |
0.34 |
| chr7_17720120_17720505 | 0.47 |
SNX13 |
sorting nexin 13 |
259779 |
0.02 |
| chr5_57134719_57134870 | 0.47 |
ENSG00000266864 |
. |
34079 |
0.24 |
| chr5_173000019_173000195 | 0.47 |
CTB-33O18.3 |
|
6539 |
0.26 |
| chr10_62335914_62336175 | 0.44 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
3623 |
0.38 |
| chr5_54456050_54456451 | 0.44 |
GPX8 |
glutathione peroxidase 8 (putative) |
252 |
0.86 |
| chr13_97930897_97931108 | 0.44 |
MBNL2 |
muscleblind-like splicing regulator 2 |
2544 |
0.4 |
| chr16_53809887_53810055 | 0.43 |
FTO |
fat mass and obesity associated |
71877 |
0.11 |
| chr11_35642709_35642958 | 0.43 |
FJX1 |
four jointed box 1 (Drosophila) |
3098 |
0.31 |
| chr3_153778293_153778444 | 0.43 |
ENSG00000243069 |
. |
20913 |
0.21 |
| chr6_134557030_134557181 | 0.43 |
ENSG00000200058 |
. |
21068 |
0.16 |
| chr8_90739151_90739302 | 0.43 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
30749 |
0.24 |
| chr6_53928552_53928746 | 0.43 |
MLIP-AS1 |
MLIP antisense RNA 1 |
15875 |
0.2 |
| chr9_16726683_16727481 | 0.42 |
RP11-62F24.2 |
|
270 |
0.85 |
| chr14_31887812_31887963 | 0.42 |
CTD-2213F21.2 |
|
1605 |
0.27 |
| chr12_118628413_118628612 | 0.42 |
TAOK3 |
TAO kinase 3 |
156 |
0.96 |
| chr15_38547811_38548278 | 0.41 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
2662 |
0.42 |
| chr14_60561615_60561766 | 0.41 |
PCNXL4 |
pecanex-like 4 (Drosophila) |
3045 |
0.25 |
| chr3_105657205_105657471 | 0.41 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
68942 |
0.15 |
| chr7_93551338_93552000 | 0.41 |
GNG11 |
guanine nucleotide binding protein (G protein), gamma 11 |
658 |
0.67 |
| chr21_40365344_40365495 | 0.41 |
ENSG00000272015 |
. |
98710 |
0.08 |
| chr3_98616068_98616219 | 0.41 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
3872 |
0.21 |
| chr8_142141679_142142152 | 0.41 |
RP11-809O17.1 |
|
1855 |
0.33 |
| chr16_28193130_28193587 | 0.41 |
XPO6 |
exportin 6 |
915 |
0.43 |
| chr9_111058733_111058884 | 0.41 |
ENSG00000222512 |
. |
62401 |
0.16 |
| chr12_50927086_50927306 | 0.40 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
28306 |
0.19 |
| chr6_27182410_27182602 | 0.40 |
PRSS16 |
protease, serine, 16 (thymus) |
32974 |
0.13 |
| chr5_107229567_107229740 | 0.40 |
ENSG00000251732 |
. |
83323 |
0.11 |
| chr1_46420407_46420664 | 0.40 |
MAST2 |
microtubule associated serine/threonine kinase 2 |
41275 |
0.16 |
| chr20_48319819_48319972 | 0.40 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
10520 |
0.19 |
| chr15_42749459_42749764 | 0.39 |
ZNF106 |
zinc finger protein 106 |
100 |
0.96 |
| chr18_67714970_67715121 | 0.39 |
RTTN |
rotatin |
27091 |
0.24 |
| chr1_173807426_173807577 | 0.39 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
13704 |
0.1 |
| chr4_122686341_122686757 | 0.39 |
TMEM155 |
transmembrane protein 155 |
33 |
0.94 |
| chr12_80081310_80081535 | 0.39 |
PAWR |
PRKC, apoptosis, WT1, regulator |
2438 |
0.38 |
| chr15_49715834_49715985 | 0.39 |
FGF7 |
fibroblast growth factor 7 |
452 |
0.86 |
| chr17_79533812_79534253 | 0.39 |
NPLOC4 |
nuclear protein localization 4 homolog (S. cerevisiae) |
371 |
0.75 |
| chr11_22724799_22724998 | 0.38 |
ENSG00000222427 |
. |
15001 |
0.2 |
| chr2_151896468_151896724 | 0.38 |
AC023469.1 |
HCG1817310; Uncharacterized protein |
8692 |
0.28 |
| chr14_101867418_101867569 | 0.38 |
ENSG00000258498 |
. |
159266 |
0.02 |
| chr2_217461840_217461991 | 0.38 |
AC073321.3 |
|
9746 |
0.19 |
| chr14_65928823_65928974 | 0.38 |
ENSG00000207781 |
. |
8922 |
0.25 |
| chr15_41316852_41317003 | 0.38 |
RP11-540O11.4 |
|
230 |
0.91 |
| chr5_15021667_15021945 | 0.38 |
ENSG00000202269 |
. |
89223 |
0.09 |
| chr8_110987229_110987457 | 0.38 |
KCNV1 |
potassium channel, subfamily V, member 1 |
384 |
0.81 |
| chr7_90012947_90013805 | 0.38 |
CLDN12 |
claudin 12 |
341 |
0.9 |
| chr6_56595678_56595829 | 0.38 |
DST |
dystonin |
54924 |
0.16 |
| chr1_234700242_234700393 | 0.37 |
ENSG00000212144 |
. |
28704 |
0.16 |
| chr18_60434411_60434742 | 0.37 |
ENSG00000206746 |
. |
36011 |
0.15 |
| chr10_101768167_101768400 | 0.37 |
DNMBP |
dynamin binding protein |
1393 |
0.42 |
| chr2_113538908_113539059 | 0.37 |
IL1A |
interleukin 1, alpha |
3184 |
0.22 |
| chr20_35923242_35923439 | 0.37 |
MANBAL |
mannosidase, beta A, lysosomal-like |
1959 |
0.35 |
| chr6_6005898_6006049 | 0.37 |
NRN1 |
neuritin 1 |
1227 |
0.56 |
| chr10_21800964_21801188 | 0.37 |
SKIDA1 |
SKI/DACH domain containing 1 |
5772 |
0.14 |
| chr1_95005421_95005705 | 0.37 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
1630 |
0.52 |
| chr6_86167890_86168219 | 0.37 |
NT5E |
5'-nucleotidase, ecto (CD73) |
8227 |
0.28 |
| chr4_44681433_44681584 | 0.37 |
YIPF7 |
Yip1 domain family, member 7 |
935 |
0.39 |
| chr6_123316934_123317301 | 0.36 |
CLVS2 |
clavesin 2 |
1 |
0.99 |
| chr18_61445658_61445809 | 0.36 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
522 |
0.82 |
| chr1_164529707_164529858 | 0.36 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
722 |
0.78 |
| chr10_17273946_17274204 | 0.36 |
VIM |
vimentin |
1467 |
0.3 |
| chr17_5420049_5420318 | 0.36 |
ENSG00000253071 |
. |
2593 |
0.22 |
| chr12_52347769_52348164 | 0.36 |
ACVR1B |
activin A receptor, type IB |
802 |
0.57 |
| chr2_9699691_9699891 | 0.36 |
ADAM17 |
ADAM metallopeptidase domain 17 |
3870 |
0.21 |
| chr15_88020923_88021173 | 0.36 |
ENSG00000207150 |
. |
3528 |
0.4 |
| chr17_49125210_49125361 | 0.36 |
SPAG9 |
sperm associated antigen 9 |
1046 |
0.5 |
| chr8_121681395_121681546 | 0.36 |
RP11-713M15.1 |
|
92023 |
0.09 |
| chr8_119427050_119427214 | 0.36 |
AC023590.1 |
Uncharacterized protein |
132651 |
0.05 |
| chr2_242257443_242257594 | 0.36 |
HDLBP |
high density lipoprotein binding protein |
1042 |
0.41 |
| chr6_148736290_148736441 | 0.36 |
SASH1 |
SAM and SH3 domain containing 1 |
72636 |
0.12 |
| chr14_62176388_62176539 | 0.35 |
HIF1A |
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
12123 |
0.24 |
| chr4_74087462_74087866 | 0.35 |
ANKRD17 |
ankyrin repeat domain 17 |
1167 |
0.53 |
| chr18_60385543_60385694 | 0.35 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
2935 |
0.28 |
| chr8_25868117_25868310 | 0.35 |
EBF2 |
early B-cell factor 2 |
30791 |
0.24 |
| chr7_97361017_97361340 | 0.35 |
TAC1 |
tachykinin, precursor 1 |
42 |
0.99 |
| chr4_26344236_26344574 | 0.35 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
357 |
0.93 |
| chr1_120255525_120255935 | 0.35 |
PHGDH |
phosphoglycerate dehydrogenase |
1220 |
0.51 |
| chr7_92224542_92224693 | 0.35 |
FAM133B |
family with sequence similarity 133, member B |
4909 |
0.24 |
| chr1_62071878_62072329 | 0.35 |
ENSG00000264551 |
. |
22600 |
0.25 |
| chr7_24988449_24988600 | 0.35 |
OSBPL3 |
oxysterol binding protein-like 3 |
30812 |
0.21 |
| chr2_55239893_55240193 | 0.35 |
ENSG00000200086 |
. |
1614 |
0.37 |
| chr9_109625885_109626257 | 0.34 |
ZNF462 |
zinc finger protein 462 |
624 |
0.77 |
| chr2_158039761_158039912 | 0.34 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
74274 |
0.11 |
| chr10_86184623_86185188 | 0.34 |
CCSER2 |
coiled-coil serine-rich protein 2 |
210 |
0.97 |
| chr21_39807445_39807798 | 0.34 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
62724 |
0.14 |
| chr3_174675589_174675952 | 0.34 |
NAALADL2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
98700 |
0.09 |
| chr17_42214986_42215221 | 0.33 |
ENSG00000212446 |
. |
1746 |
0.18 |
| chr7_70201396_70202343 | 0.33 |
AUTS2 |
autism susceptibility candidate 2 |
7744 |
0.34 |
| chr16_55401499_55401650 | 0.33 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
22038 |
0.18 |
| chr4_148947037_148947237 | 0.33 |
RP11-76G10.1 |
|
120485 |
0.06 |
| chr5_142562344_142562495 | 0.33 |
ARHGAP26-IT1 |
ARHGAP26 intronic transcript 1 (non-protein coding) |
9646 |
0.25 |
| chr11_131780111_131780399 | 0.33 |
NTM |
neurotrimin |
642 |
0.77 |
| chr5_16593515_16593666 | 0.33 |
RP11-260E18.1 |
|
22445 |
0.17 |
| chr1_45083000_45083241 | 0.33 |
RNF220 |
ring finger protein 220 |
8878 |
0.17 |
| chr3_54897831_54897982 | 0.33 |
CACNA2D3-AS1 |
CACNA2D3 antisense RNA 1 |
37376 |
0.21 |
| chr3_11556089_11556255 | 0.33 |
VGLL4 |
vestigial like 4 (Drosophila) |
54226 |
0.15 |
| chr12_49581087_49581445 | 0.33 |
TUBA1A |
tubulin, alpha 1a |
25 |
0.96 |
| chr2_109210392_109210708 | 0.33 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
5628 |
0.26 |
| chr2_176992012_176992333 | 0.32 |
HOXD8 |
homeobox D8 |
2250 |
0.12 |
| chr7_39872474_39872794 | 0.32 |
ENSG00000242855 |
. |
55338 |
0.13 |
| chr12_109122720_109122871 | 0.32 |
CORO1C |
coronin, actin binding protein, 1C |
1532 |
0.34 |
| chr12_10548505_10548720 | 0.32 |
KLRK1 |
killer cell lectin-like receptor subfamily K, member 1 |
5995 |
0.13 |
| chr3_88191159_88191414 | 0.32 |
ZNF654 |
zinc finger protein 654 |
3032 |
0.24 |
| chr12_41224022_41224173 | 0.32 |
CNTN1 |
contactin 1 |
2111 |
0.48 |
| chr8_87317262_87317629 | 0.32 |
WWP1 |
WW domain containing E3 ubiquitin protein ligase 1 |
37522 |
0.18 |
| chr5_19885020_19885171 | 0.31 |
CDH18 |
cadherin 18, type 2 |
1286 |
0.64 |
| chr1_102791433_102791584 | 0.31 |
OLFM3 |
olfactomedin 3 |
328922 |
0.01 |
| chr2_159456331_159456527 | 0.31 |
ENSG00000251721 |
. |
65717 |
0.12 |
| chr2_239695781_239695932 | 0.31 |
TWIST2 |
twist family bHLH transcription factor 2 |
60817 |
0.13 |
| chr18_28779723_28779874 | 0.31 |
DSC1 |
desmocollin 1 |
36979 |
0.15 |
| chr5_38807697_38807893 | 0.31 |
RP11-122C5.3 |
|
24113 |
0.22 |
| chr4_48779368_48779519 | 0.31 |
FRYL |
FRY-like |
2822 |
0.3 |
| chr8_126231853_126232267 | 0.31 |
ENSG00000242170 |
. |
50786 |
0.15 |
| chr18_9742659_9743173 | 0.31 |
RAB31 |
RAB31, member RAS oncogene family |
34754 |
0.17 |
| chr13_86159310_86159559 | 0.31 |
ENSG00000207012 |
. |
446 |
0.91 |
| chr4_177824622_177824937 | 0.31 |
VEGFC |
vascular endothelial growth factor C |
110898 |
0.07 |
| chr2_161224712_161224959 | 0.31 |
ENSG00000252465 |
. |
28644 |
0.18 |
| chr4_42361739_42361897 | 0.31 |
RP11-63A11.1 |
|
31432 |
0.19 |
| chr9_75568093_75568244 | 0.31 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
196 |
0.97 |
| chr2_55810269_55810420 | 0.31 |
ENSG00000212175 |
. |
17505 |
0.15 |
| chr14_71838887_71839322 | 0.30 |
ENSG00000207444 |
. |
25950 |
0.19 |
| chr2_207998546_207999225 | 0.30 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
24 |
0.98 |
| chr10_78157088_78157239 | 0.30 |
RP11-369F10.2 |
|
41370 |
0.18 |
| chr4_178222633_178222784 | 0.30 |
NEIL3 |
nei endonuclease VIII-like 3 (E. coli) |
8282 |
0.23 |
| chr16_88472278_88472669 | 0.30 |
ZNF469 |
zinc finger protein 469 |
21406 |
0.18 |
| chr8_99334814_99335068 | 0.30 |
NIPAL2 |
NIPA-like domain containing 2 |
28320 |
0.2 |
| chr17_67137246_67137397 | 0.30 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
694 |
0.69 |
| chr15_63050120_63050513 | 0.30 |
TLN2 |
talin 2 |
473 |
0.82 |
| chr10_5491792_5492244 | 0.30 |
NET1 |
neuroepithelial cell transforming 1 |
3444 |
0.22 |
| chr13_23946605_23946801 | 0.30 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
2979 |
0.39 |
| chr1_100317573_100317724 | 0.30 |
AGL |
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
578 |
0.74 |
| chr11_111848673_111848997 | 0.30 |
DIXDC1 |
DIX domain containing 1 |
802 |
0.5 |
| chr13_106938318_106938469 | 0.30 |
ENSG00000222682 |
. |
130554 |
0.06 |
| chr3_114477857_114478359 | 0.30 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
10 |
0.99 |
| chr14_96723158_96723326 | 0.30 |
BDKRB1 |
bradykinin receptor B1 |
695 |
0.57 |
| chr5_62570567_62570718 | 0.30 |
HTR1A |
5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled |
687138 |
0.0 |
| chr16_51671642_51672070 | 0.29 |
ENSG00000223168 |
. |
33618 |
0.25 |
| chr5_102000842_102000993 | 0.29 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
88768 |
0.1 |
| chr13_73631347_73631498 | 0.29 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
1508 |
0.47 |
| chr14_52339728_52339879 | 0.29 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
4507 |
0.23 |
| chr8_29027576_29027748 | 0.29 |
ENSG00000264328 |
. |
27452 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.2 | 0.6 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.2 | 0.8 | GO:0060437 | lung growth(GO:0060437) |
| 0.2 | 0.5 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.3 | GO:0031394 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 0.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.2 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.1 | 0.2 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 0.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.1 | GO:0002246 | wound healing involved in inflammatory response(GO:0002246) inflammatory response to wounding(GO:0090594) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.1 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.1 | 0.3 | GO:0010664 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.4 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 0.6 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.1 | 0.2 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.1 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) |
| 0.1 | 0.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.2 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.2 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.2 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.4 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:1903170 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.2 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0060526 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.1 | GO:0006623 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.2 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.2 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.0 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.0 | GO:1900078 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.0 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.0 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.1 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.0 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.0 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0090224 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 0.0 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 1.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.5 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.0 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.1 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |