Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
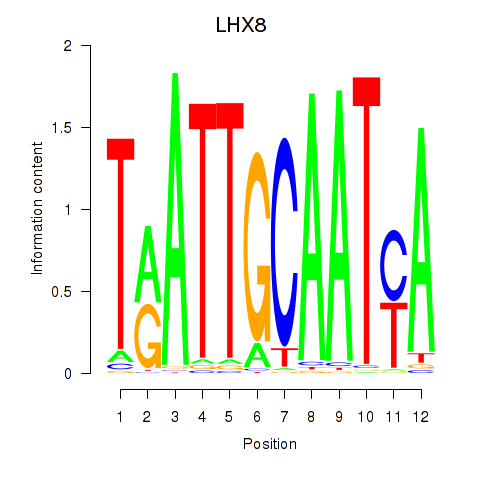
Results for LHX8
Z-value: 1.13
Transcription factors associated with LHX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX8
|
ENSG00000162624.10 | LIM homeobox 8 |
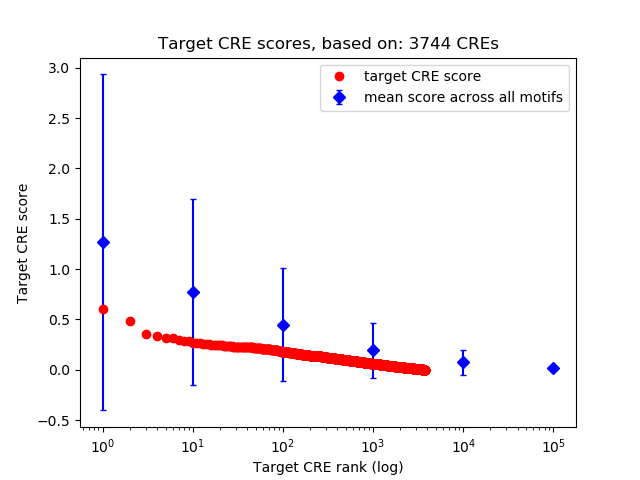
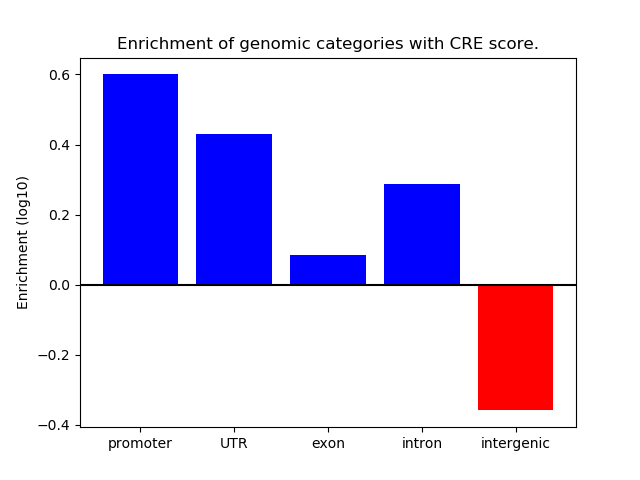
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_75571987_75572138 | LHX8 | 22057 | 0.172584 | 0.22 | 5.8e-01 | Click! |
| chr1_75600688_75601584 | LHX8 | 569 | 0.698057 | -0.21 | 5.9e-01 | Click! |
Activity of the LHX8 motif across conditions
Conditions sorted by the z-value of the LHX8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
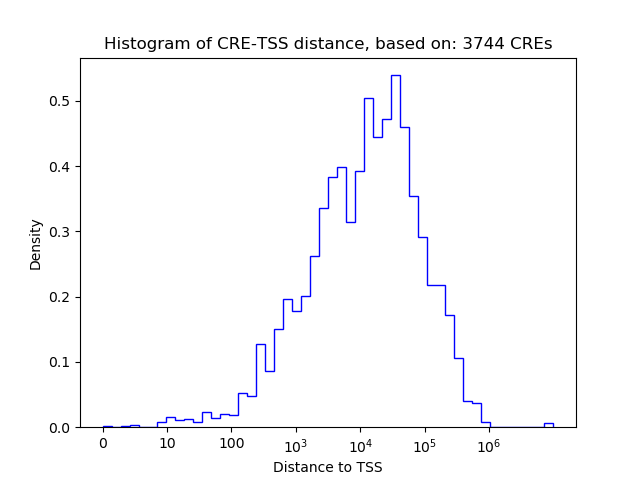
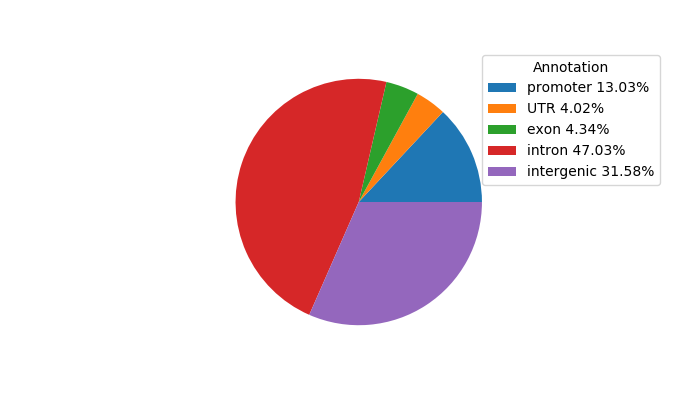
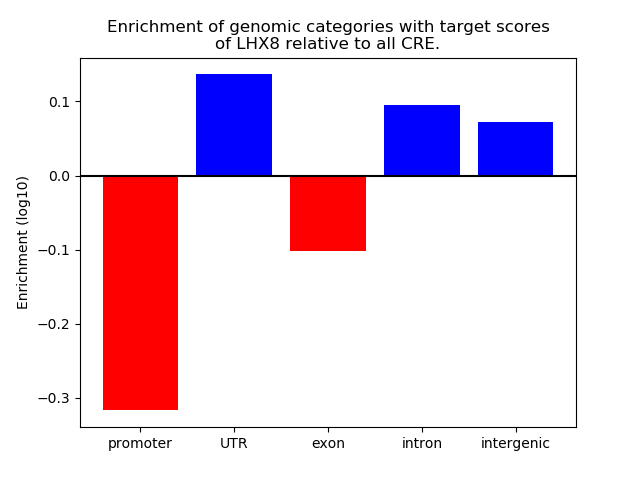
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_28373361_28373512 | 0.60 |
LINGO2 |
leucine rich repeat and Ig domain containing 2 |
296847 |
0.01 |
| chr7_76785964_76786115 | 0.49 |
CCDC146 |
coiled-coil domain containing 146 |
34105 |
0.15 |
| chr18_72647452_72647603 | 0.36 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
269941 |
0.02 |
| chr1_227501047_227501198 | 0.34 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
3761 |
0.35 |
| chr8_66868091_66868242 | 0.32 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
65629 |
0.13 |
| chr2_233565064_233565215 | 0.31 |
GIGYF2 |
GRB10 interacting GYF protein 2 |
3077 |
0.19 |
| chr4_39570973_39571124 | 0.29 |
UGDH |
UDP-glucose 6-dehydrogenase |
41117 |
0.11 |
| chr10_4023404_4023555 | 0.29 |
KLF6 |
Kruppel-like factor 6 |
196006 |
0.03 |
| chr1_66806017_66806168 | 0.29 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
8220 |
0.31 |
| chr22_17598033_17598230 | 0.27 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
4012 |
0.17 |
| chr22_41249705_41249922 | 0.26 |
ST13 |
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
789 |
0.51 |
| chrX_68835519_68835670 | 0.26 |
EDA |
ectodysplasin A |
317 |
0.92 |
| chr18_30563491_30563642 | 0.26 |
RP11-680N20.1 |
|
29527 |
0.24 |
| chr10_27312512_27312663 | 0.25 |
ANKRD26 |
ankyrin repeat domain 26 |
8979 |
0.23 |
| chr11_122502980_122503131 | 0.25 |
UBASH3B |
ubiquitin associated and SH3 domain containing B |
23328 |
0.21 |
| chr2_55986068_55986219 | 0.25 |
PNPT1 |
polyribonucleotide nucleotidyltransferase 1 |
65098 |
0.11 |
| chr9_3827440_3827591 | 0.25 |
RP11-252M18.3 |
|
48069 |
0.17 |
| chr2_231429944_231430095 | 0.25 |
AC010149.4 |
|
14629 |
0.18 |
| chr6_10522784_10522935 | 0.25 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
1275 |
0.45 |
| chr8_90031831_90031982 | 0.24 |
RP11-586K2.1 |
|
319386 |
0.01 |
| chr7_38400564_38400715 | 0.24 |
AMPH |
amphiphysin |
102074 |
0.08 |
| chr10_89692060_89692211 | 0.24 |
RP11-380G5.2 |
|
53686 |
0.11 |
| chr7_116341288_116341439 | 0.24 |
MET |
met proto-oncogene |
2224 |
0.39 |
| chr5_158301830_158301981 | 0.23 |
CTD-2363C16.1 |
|
108109 |
0.07 |
| chr18_12596428_12596579 | 0.23 |
SPIRE1 |
spire-type actin nucleation factor 1 |
56581 |
0.09 |
| chr14_105759141_105759292 | 0.23 |
BRF1 |
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
7589 |
0.16 |
| chr20_52726952_52727103 | 0.23 |
BCAS1 |
breast carcinoma amplified sequence 1 |
39723 |
0.15 |
| chr13_78109984_78110135 | 0.23 |
SCEL |
sciellin |
164 |
0.97 |
| chr2_242063252_242063403 | 0.23 |
PASK |
PAS domain containing serine/threonine kinase |
15683 |
0.13 |
| chr14_52867726_52867877 | 0.23 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
86688 |
0.09 |
| chr2_200778375_200778671 | 0.23 |
C2orf69 |
chromosome 2 open reading frame 69 |
2544 |
0.27 |
| chrX_53253512_53254652 | 0.23 |
KDM5C |
lysine (K)-specific demethylase 5C |
276 |
0.9 |
| chr9_99265682_99265833 | 0.23 |
HABP4 |
hyaluronan binding protein 4 |
53270 |
0.13 |
| chr2_196548060_196548211 | 0.23 |
ENSG00000201813 |
. |
12937 |
0.23 |
| chr9_128411357_128411651 | 0.23 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
1192 |
0.6 |
| chr9_112813674_112813882 | 0.23 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
2802 |
0.39 |
| chr5_77224879_77225030 | 0.22 |
TBCA |
tubulin folding cofactor A |
60350 |
0.14 |
| chr7_115670465_115670618 | 0.22 |
TFEC |
transcription factor EC |
254 |
0.96 |
| chr11_75535232_75535410 | 0.22 |
RP11-535A19.2 |
|
8870 |
0.1 |
| chr4_146763857_146764008 | 0.22 |
RP11-181K12.2 |
|
9662 |
0.26 |
| chr4_38162963_38163114 | 0.22 |
ENSG00000221495 |
. |
77196 |
0.11 |
| chr7_37755543_37755694 | 0.22 |
GPR141 |
G protein-coupled receptor 141 |
24378 |
0.21 |
| chr9_132708790_132708941 | 0.22 |
RP11-409K20.6 |
|
13050 |
0.19 |
| chr5_127688862_127689013 | 0.22 |
FBN2 |
fibrillin 2 |
14054 |
0.24 |
| chr6_124159912_124160063 | 0.22 |
RP11-374A22.1 |
|
15602 |
0.24 |
| chr5_67562966_67563283 | 0.22 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
12944 |
0.27 |
| chr1_114517596_114517747 | 0.22 |
OLFML3 |
olfactomedin-like 3 |
4392 |
0.19 |
| chr4_48701192_48701343 | 0.22 |
FRYL |
FRY-like |
18079 |
0.24 |
| chr3_185445450_185445601 | 0.22 |
C3orf65 |
chromosome 3 open reading frame 65 |
14445 |
0.22 |
| chr10_90754998_90755312 | 0.22 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
4008 |
0.17 |
| chr13_100225293_100225444 | 0.22 |
CLYBL |
citrate lyase beta like |
33551 |
0.14 |
| chr6_106500939_106501090 | 0.22 |
PRDM1 |
PR domain containing 1, with ZNF domain |
33181 |
0.19 |
| chr11_75584822_75585009 | 0.22 |
UVRAG |
UV radiation resistance associated |
4533 |
0.16 |
| chr4_56501775_56502572 | 0.22 |
NMU |
neuromedin U |
253 |
0.93 |
| chr9_21024320_21024568 | 0.21 |
PTPLAD2 |
protein tyrosine phosphatase-like A domain containing 2 |
7164 |
0.22 |
| chr19_10697425_10697576 | 0.21 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
395 |
0.72 |
| chr8_127513113_127513264 | 0.21 |
ENSG00000207138 |
. |
3820 |
0.28 |
| chr10_3807723_3807874 | 0.21 |
RP11-184A2.2 |
|
2309 |
0.32 |
| chr13_70839300_70839451 | 0.21 |
KLHL1 |
kelch-like family member 1 |
156784 |
0.04 |
| chr9_123958868_123959019 | 0.21 |
RAB14 |
RAB14, member RAS oncogene family |
5204 |
0.17 |
| chr2_101197927_101198078 | 0.21 |
ENSG00000238328 |
. |
4071 |
0.2 |
| chr7_38903515_38903666 | 0.21 |
VPS41 |
vacuolar protein sorting 41 homolog (S. cerevisiae) |
32644 |
0.22 |
| chr6_111888477_111888628 | 0.21 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
11 |
0.97 |
| chr11_65886976_65887127 | 0.21 |
PACS1 |
phosphofurin acidic cluster sorting protein 1 |
18817 |
0.1 |
| chrX_22863600_22863751 | 0.21 |
DDX53 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 53 |
154412 |
0.04 |
| chr9_687749_687900 | 0.21 |
RP11-130C19.3 |
|
2269 |
0.34 |
| chr17_56477664_56477815 | 0.21 |
RNF43 |
ring finger protein 43 |
2343 |
0.23 |
| chr7_41478323_41478624 | 0.21 |
INHBA-AS1 |
INHBA antisense RNA 1 |
255041 |
0.02 |
| chr19_8922272_8922597 | 0.20 |
CTD-2529P6.3 |
|
9743 |
0.14 |
| chr2_173995675_173995826 | 0.20 |
MLTK |
Mitogen-activated protein kinase kinase kinase MLT |
40403 |
0.17 |
| chr4_39033572_39034012 | 0.20 |
TMEM156 |
transmembrane protein 156 |
249 |
0.93 |
| chr11_63915867_63916232 | 0.20 |
MACROD1 |
MACRO domain containing 1 |
17484 |
0.09 |
| chr2_153323935_153324293 | 0.20 |
FMNL2 |
formin-like 2 |
132363 |
0.05 |
| chr11_105890749_105891017 | 0.20 |
MSANTD4 |
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
2071 |
0.32 |
| chr13_30168298_30169638 | 0.20 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
857 |
0.74 |
| chr17_40739225_40739376 | 0.20 |
PSMC3IP |
PSMC3 interacting protein |
9451 |
0.08 |
| chr22_40856327_40856478 | 0.20 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
3020 |
0.24 |
| chr15_59646896_59647047 | 0.20 |
RP11-356M20.3 |
|
5117 |
0.14 |
| chr10_45123233_45123384 | 0.20 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
242768 |
0.02 |
| chr13_30947300_30947451 | 0.20 |
KATNAL1 |
katanin p60 subunit A-like 1 |
65754 |
0.12 |
| chrX_13007277_13007565 | 0.20 |
TMSB4X |
thymosin beta 4, X-linked |
13644 |
0.22 |
| chr15_33135515_33135666 | 0.19 |
FMN1 |
formin 1 |
44865 |
0.14 |
| chr4_143252459_143252732 | 0.19 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
25498 |
0.28 |
| chr5_24644888_24645378 | 0.19 |
CDH10 |
cadherin 10, type 2 (T2-cadherin) |
46 |
0.99 |
| chr10_53221961_53222112 | 0.19 |
RP11-539E19.2 |
|
159710 |
0.04 |
| chr20_39135469_39135620 | 0.19 |
ENSG00000252434 |
. |
90029 |
0.1 |
| chr1_243420909_243421197 | 0.19 |
SDCCAG8 |
serologically defined colon cancer antigen 8 |
1681 |
0.33 |
| chr10_76689232_76689383 | 0.19 |
KAT6B |
K(lysine) acetyltransferase 6B |
90849 |
0.07 |
| chrX_131341527_131341678 | 0.19 |
RAP2C-AS1 |
RAP2C antisense RNA 1 |
9573 |
0.22 |
| chr2_151124427_151124578 | 0.19 |
RND3 |
Rho family GTPase 3 |
217394 |
0.02 |
| chr9_125798505_125798656 | 0.19 |
GPR21 |
G protein-coupled receptor 21 |
1774 |
0.31 |
| chr6_157129169_157129320 | 0.19 |
RP11-230C9.3 |
|
27655 |
0.18 |
| chr2_208602192_208602343 | 0.19 |
CCNYL1 |
cyclin Y-like 1 |
4731 |
0.16 |
| chr4_123588404_123588555 | 0.18 |
IL21 |
interleukin 21 |
46255 |
0.14 |
| chr4_71891789_71891940 | 0.18 |
DCK |
deoxycytidine kinase |
32509 |
0.21 |
| chr12_94161563_94161714 | 0.18 |
RP11-887P2.5 |
|
30039 |
0.17 |
| chr9_16679077_16679228 | 0.18 |
BNC2 |
basonuclin 2 |
25928 |
0.2 |
| chr7_80704969_80705120 | 0.18 |
AC005008.2 |
Uncharacterized protein |
99780 |
0.09 |
| chr18_51736710_51736861 | 0.18 |
ENSG00000207233 |
. |
11997 |
0.21 |
| chr5_131014671_131014822 | 0.18 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
43817 |
0.18 |
| chr12_10558293_10558444 | 0.18 |
KLRC4 |
killer cell lectin-like receptor subfamily C, member 4 |
3988 |
0.14 |
| chr12_95347508_95347659 | 0.18 |
NDUFA12 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
49893 |
0.15 |
| chr14_25147155_25147306 | 0.18 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
43757 |
0.13 |
| chr1_8360871_8361022 | 0.18 |
ENSG00000251977 |
. |
8623 |
0.16 |
| chr6_25968730_25968889 | 0.18 |
TRIM38 |
tripartite motif containing 38 |
5534 |
0.11 |
| chr9_111075511_111075662 | 0.18 |
ENSG00000222512 |
. |
45623 |
0.21 |
| chr15_57520158_57520309 | 0.18 |
TCF12 |
transcription factor 12 |
8569 |
0.27 |
| chr7_93366478_93366629 | 0.18 |
ENSG00000265423 |
. |
20313 |
0.24 |
| chr3_9745623_9746706 | 0.18 |
CPNE9 |
copine family member IX |
654 |
0.62 |
| chr5_75545047_75545198 | 0.18 |
RP11-466P24.6 |
|
62165 |
0.14 |
| chr4_111821276_111821427 | 0.18 |
ENSG00000215961 |
. |
39548 |
0.19 |
| chrX_70293235_70293425 | 0.18 |
SNX12 |
sorting nexin 12 |
5057 |
0.13 |
| chr11_3923942_3924093 | 0.17 |
STIM1 |
stromal interaction molecule 1 |
676 |
0.62 |
| chr10_63595750_63596410 | 0.17 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
64979 |
0.13 |
| chr5_158445903_158446054 | 0.17 |
CTD-2363C16.2 |
|
33164 |
0.18 |
| chr7_12729528_12729679 | 0.17 |
ARL4A |
ADP-ribosylation factor-like 4A |
2343 |
0.34 |
| chr5_29222993_29223144 | 0.17 |
ENSG00000252601 |
. |
152465 |
0.05 |
| chr9_88953667_88953818 | 0.17 |
ZCCHC6 |
zinc finger, CCHC domain containing 6 |
4296 |
0.27 |
| chr7_15720473_15720624 | 0.17 |
MEOX2 |
mesenchyme homeobox 2 |
5889 |
0.26 |
| chr12_107774173_107774467 | 0.17 |
ENSG00000200897 |
. |
5796 |
0.28 |
| chr4_40516329_40516480 | 0.17 |
RBM47 |
RNA binding motif protein 47 |
156 |
0.96 |
| chr9_81760516_81760667 | 0.17 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
426097 |
0.01 |
| chr1_39329527_39329678 | 0.17 |
RP5-864K19.4 |
|
3847 |
0.16 |
| chr4_36245078_36245884 | 0.17 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
80 |
0.74 |
| chr5_54127651_54127802 | 0.17 |
ENSG00000221073 |
. |
21050 |
0.24 |
| chr10_122451233_122451392 | 0.17 |
WDR11-AS1 |
WDR11 antisense RNA 1 |
85084 |
0.09 |
| chr4_124593202_124593353 | 0.17 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
272154 |
0.02 |
| chr14_38889256_38889407 | 0.17 |
CLEC14A |
C-type lectin domain family 14, member A |
163757 |
0.04 |
| chr11_36618151_36618302 | 0.17 |
RAG2 |
recombination activating gene 2 |
1560 |
0.34 |
| chr13_108457167_108457318 | 0.17 |
FAM155A-IT1 |
FAM155A intronic transcript 1 (non-protein coding) |
30557 |
0.22 |
| chr18_56270622_56270891 | 0.17 |
ENSG00000252284 |
. |
2893 |
0.22 |
| chr2_109539318_109539469 | 0.17 |
CCDC138 |
coiled-coil domain containing 138 |
66009 |
0.12 |
| chr2_102357284_102357468 | 0.17 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
42384 |
0.2 |
| chr4_95562323_95562474 | 0.17 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
116721 |
0.07 |
| chr2_213976639_213976790 | 0.17 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
36639 |
0.21 |
| chr11_10826698_10826955 | 0.16 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
2183 |
0.2 |
| chr19_45682453_45683430 | 0.16 |
BLOC1S3 |
biogenesis of lysosomal organelles complex-1, subunit 3 |
14 |
0.95 |
| chr21_39847210_39847361 | 0.16 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
23060 |
0.27 |
| chr1_195877260_195877411 | 0.16 |
ENSG00000265986 |
. |
674276 |
0.0 |
| chr7_6732804_6732968 | 0.16 |
ENSG00000265245 |
. |
1278 |
0.34 |
| chr5_17222098_17222777 | 0.16 |
BASP1 |
brain abundant, membrane attached signal protein 1 |
4768 |
0.2 |
| chr5_114193091_114193242 | 0.16 |
RP11-492A10.1 |
|
189059 |
0.03 |
| chr14_64804121_64804291 | 0.16 |
ESR2 |
estrogen receptor 2 (ER beta) |
608 |
0.56 |
| chr14_31116263_31116414 | 0.16 |
SCFD1 |
sec1 family domain containing 1 |
7355 |
0.25 |
| chr6_157381110_157381404 | 0.16 |
RP1-137K2.2 |
|
59583 |
0.15 |
| chr17_48393570_48393721 | 0.16 |
RP11-893F2.9 |
|
28429 |
0.09 |
| chr2_74802682_74802833 | 0.16 |
LOXL3 |
lysyl oxidase-like 3 |
19940 |
0.09 |
| chr21_27500577_27500761 | 0.16 |
APP |
amyloid beta (A4) precursor protein |
12109 |
0.22 |
| chr16_79731835_79731986 | 0.16 |
ENSG00000221330 |
. |
28276 |
0.25 |
| chr3_107937522_107937673 | 0.16 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
3625 |
0.33 |
| chr5_53912070_53912221 | 0.16 |
SNX18 |
sorting nexin 18 |
98552 |
0.08 |
| chr5_131991447_131991948 | 0.16 |
AC004041.2 |
|
113 |
0.94 |
| chr3_5343632_5343783 | 0.15 |
ENSG00000241227 |
. |
48799 |
0.14 |
| chrX_114276029_114276180 | 0.15 |
ENSG00000222122 |
. |
9114 |
0.21 |
| chr10_31603702_31603954 | 0.15 |
ENSG00000237036 |
. |
4033 |
0.23 |
| chr18_56282753_56282991 | 0.15 |
ALPK2 |
alpha-kinase 2 |
13317 |
0.15 |
| chr4_164416977_164417128 | 0.15 |
TMA16 |
translation machinery associated 16 homolog (S. cerevisiae) |
1192 |
0.5 |
| chrX_64901585_64901736 | 0.15 |
MSN |
moesin |
14123 |
0.3 |
| chr13_80441817_80441968 | 0.15 |
NDFIP2 |
Nedd4 family interacting protein 2 |
386304 |
0.01 |
| chr2_178649432_178649708 | 0.15 |
AC012499.1 |
|
86352 |
0.08 |
| chr18_63233244_63233395 | 0.15 |
CDH7 |
cadherin 7, type 2 |
184169 |
0.03 |
| chr10_33240556_33240863 | 0.15 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
3882 |
0.33 |
| chr7_116416437_116416588 | 0.15 |
MET |
met proto-oncogene |
717 |
0.73 |
| chr2_177679359_177679510 | 0.15 |
ENSG00000206866 |
. |
115384 |
0.06 |
| chr6_152451471_152451622 | 0.15 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
37940 |
0.22 |
| chr1_21276828_21277022 | 0.15 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
8814 |
0.23 |
| chr16_12667643_12667794 | 0.15 |
CTD-3037G24.4 |
|
16167 |
0.18 |
| chr3_112183991_112184142 | 0.15 |
BTLA |
B and T lymphocyte associated |
34139 |
0.19 |
| chr1_98853949_98854194 | 0.15 |
ENSG00000221777 |
. |
15130 |
0.31 |
| chr20_13903186_13903337 | 0.15 |
ENSG00000221328 |
. |
33242 |
0.17 |
| chr15_80365779_80365947 | 0.15 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
931 |
0.64 |
| chr6_120831058_120831209 | 0.15 |
ENSG00000206857 |
. |
16413 |
0.3 |
| chr7_84267660_84267811 | 0.15 |
ENSG00000265050 |
. |
27589 |
0.27 |
| chr13_30415572_30415723 | 0.15 |
UBL3 |
ubiquitin-like 3 |
9174 |
0.3 |
| chr15_70728001_70728152 | 0.15 |
ENSG00000200216 |
. |
242501 |
0.02 |
| chr2_66668625_66669005 | 0.15 |
AC092669.1 |
|
153 |
0.89 |
| chr2_167042360_167042511 | 0.15 |
ENSG00000222376 |
. |
4959 |
0.27 |
| chr1_180396421_180396572 | 0.15 |
ENSG00000265435 |
. |
11029 |
0.24 |
| chr2_40611863_40612014 | 0.15 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
45482 |
0.19 |
| chr4_114758520_114758671 | 0.15 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
75512 |
0.11 |
| chr5_57787974_57788125 | 0.15 |
GAPT |
GRB2-binding adaptor protein, transmembrane |
785 |
0.66 |
| chr8_115880767_115880918 | 0.15 |
TRPS1 |
trichorhinophalangeal syndrome I |
623606 |
0.0 |
| chr2_158643187_158643338 | 0.15 |
ACVR1 |
activin A receptor, type I |
20246 |
0.22 |
| chr18_59617014_59617243 | 0.15 |
RNF152 |
ring finger protein 152 |
55664 |
0.17 |
| chr16_48325554_48325705 | 0.15 |
LONP2 |
lon peptidase 2, peroxisomal |
42594 |
0.14 |
| chr18_22925102_22925253 | 0.15 |
ZNF521 |
zinc finger protein 521 |
5980 |
0.3 |
| chr17_30878109_30878260 | 0.15 |
RP11-466A19.5 |
|
21117 |
0.12 |
| chr8_134512578_134512847 | 0.15 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
1086 |
0.67 |
| chr9_10292066_10292217 | 0.14 |
PTPRD |
protein tyrosine phosphatase, receptor type, D |
258351 |
0.02 |
| chr10_75668708_75669249 | 0.14 |
PLAU |
plasminogen activator, urokinase |
43 |
0.97 |
| chr6_106962020_106962173 | 0.14 |
AIM1 |
absent in melanoma 1 |
2366 |
0.31 |
| chr1_34368788_34368939 | 0.14 |
RP5-1007G16.1 |
|
27796 |
0.2 |
| chr6_148650626_148650914 | 0.14 |
SASH1 |
SAM and SH3 domain containing 1 |
12959 |
0.24 |
| chr18_63063126_63063277 | 0.14 |
CDH7 |
cadherin 7, type 2 |
354287 |
0.01 |
| chr4_178235041_178235192 | 0.14 |
NEIL3 |
nei endonuclease VIII-like 3 (E. coli) |
4126 |
0.26 |
| chr4_26198805_26199180 | 0.14 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
33915 |
0.24 |
| chr2_64320008_64320159 | 0.14 |
AC074289.1 |
|
50290 |
0.14 |
| chr15_59223516_59223667 | 0.14 |
SLTM |
SAFB-like, transcription modulator |
2182 |
0.3 |
| chr1_160678676_160678904 | 0.14 |
CD48 |
CD48 molecule |
2803 |
0.22 |
| chr6_138084606_138084757 | 0.14 |
ENSG00000207300 |
. |
20830 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.2 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.3 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.1 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0006925 | inflammatory cell apoptotic process(GO:0006925) |
| 0.0 | 0.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.3 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:0046398 | UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:0071732 | cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.1 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0031579 | membrane raft organization(GO:0031579) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.0 | GO:0001711 | endodermal cell fate commitment(GO:0001711) endodermal cell fate specification(GO:0001714) endodermal cell differentiation(GO:0035987) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031082 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0018995 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |