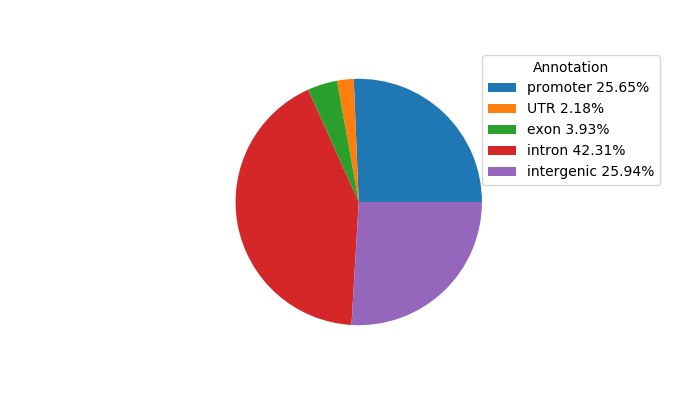
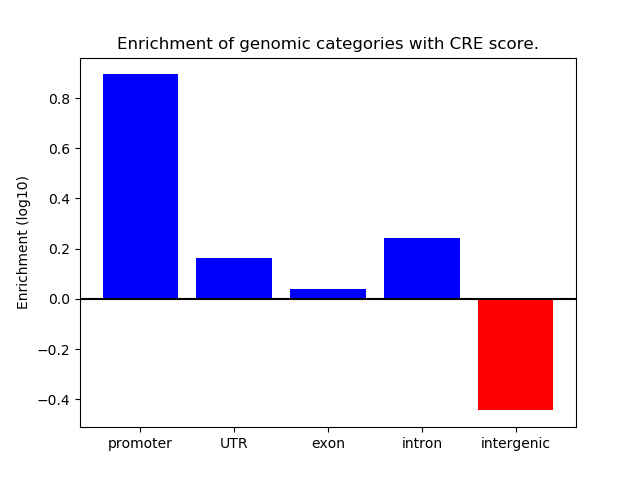
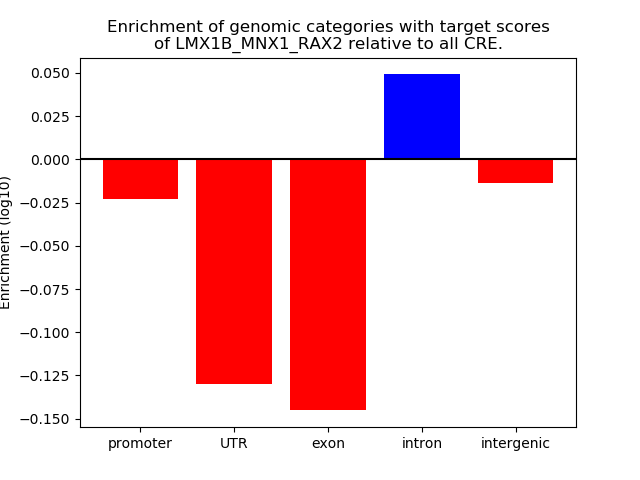
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for LMX1B_MNX1_RAX2
Z-value: 0.53
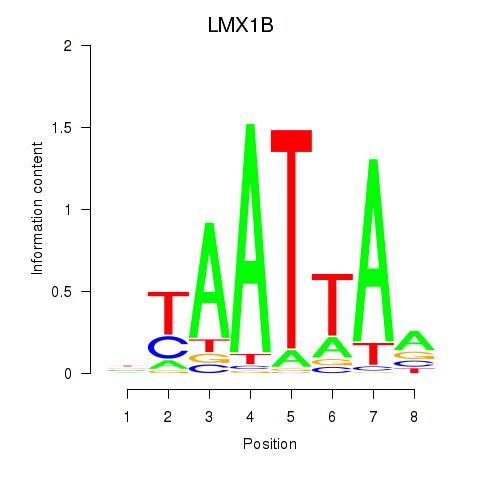
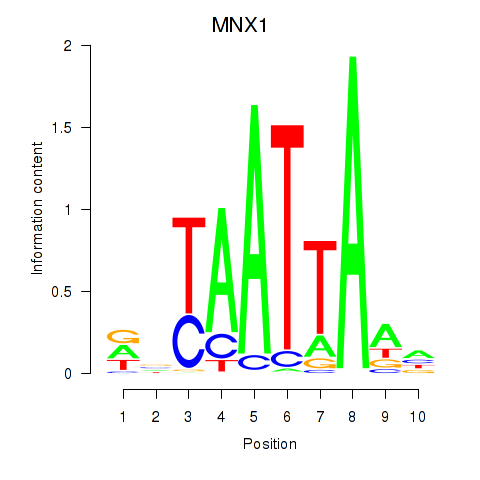
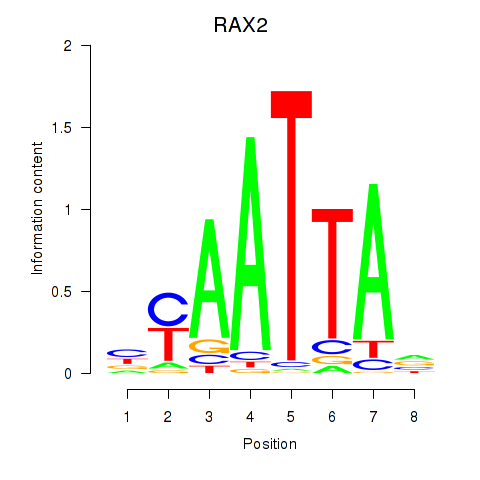
Transcription factors associated with LMX1B_MNX1_RAX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LMX1B
|
ENSG00000136944.13 | LIM homeobox transcription factor 1 beta |
|
MNX1
|
ENSG00000130675.10 | motor neuron and pancreas homeobox 1 |
|
RAX2
|
ENSG00000173976.11 | retina and anterior neural fold homeobox 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_129377175_129377421 | LMX1B | 550 | 0.655103 | 0.44 | 2.4e-01 | Click! |
| chr9_129388182_129388507 | LMX1B | 11596 | 0.158307 | 0.24 | 5.4e-01 | Click! |
| chr9_129388558_129388709 | LMX1B | 11885 | 0.157836 | -0.09 | 8.2e-01 | Click! |
| chr9_129376782_129376983 | LMX1B | 134 | 0.913436 | 0.07 | 8.5e-01 | Click! |
| chr7_156802935_156803420 | MNX1 | 168 | 0.685714 | 0.53 | 1.4e-01 | Click! |
| chr7_156802235_156802413 | MNX1 | 67 | 0.950026 | 0.53 | 1.4e-01 | Click! |
| chr7_156801299_156801450 | MNX1 | 395 | 0.789112 | 0.42 | 2.6e-01 | Click! |
| chr7_156801968_156802119 | MNX1 | 62 | 0.956588 | 0.21 | 6.0e-01 | Click! |
Activity of the LMX1B_MNX1_RAX2 motif across conditions
Conditions sorted by the z-value of the LMX1B_MNX1_RAX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
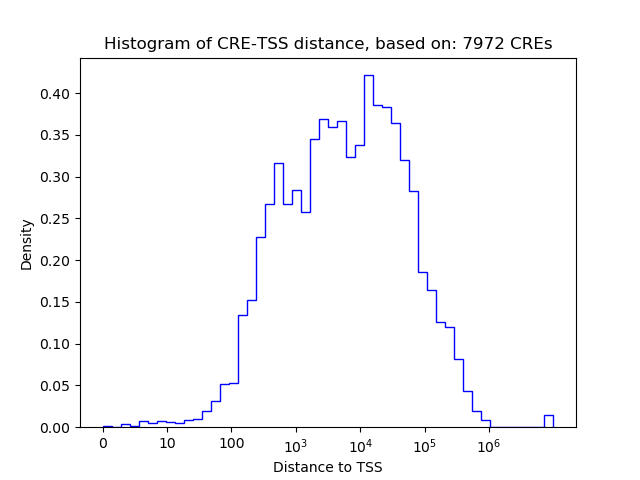
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_91881184_91881623 | 0.24 |
CCDC88C |
coiled-coil domain containing 88C |
2287 |
0.37 |
| chr6_159071306_159071615 | 0.24 |
SYTL3 |
synaptotagmin-like 3 |
414 |
0.83 |
| chr2_213962198_213962349 | 0.24 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
51080 |
0.16 |
| chr16_71935673_71935824 | 0.23 |
IST1 |
increased sodium tolerance 1 homolog (yeast) |
6256 |
0.13 |
| chr10_33425652_33425891 | 0.22 |
ENSG00000263576 |
. |
38207 |
0.16 |
| chr2_204721096_204721247 | 0.22 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
11338 |
0.25 |
| chr12_4385579_4385730 | 0.21 |
CCND2-AS1 |
CCND2 antisense RNA 1 |
304 |
0.58 |
| chr1_113164228_113164419 | 0.21 |
ST7L |
suppression of tumorigenicity 7 like |
2283 |
0.19 |
| chr10_677138_677289 | 0.20 |
RP11-809C18.3 |
|
2635 |
0.23 |
| chr9_92032170_92032321 | 0.20 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
1503 |
0.49 |
| chr14_35306917_35307068 | 0.19 |
ENSG00000251726 |
. |
7574 |
0.17 |
| chr14_22968352_22968523 | 0.19 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
12266 |
0.1 |
| chrX_19815653_19816006 | 0.19 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
2040 |
0.46 |
| chr3_168442261_168442412 | 0.19 |
ENSG00000207717 |
. |
172694 |
0.04 |
| chr1_226251298_226251924 | 0.19 |
H3F3A |
H3 histone, family 3A |
67 |
0.97 |
| chr1_180387724_180387875 | 0.19 |
ENSG00000265435 |
. |
19726 |
0.22 |
| chr1_192546230_192546423 | 0.18 |
RGS1 |
regulator of G-protein signaling 1 |
1423 |
0.45 |
| chr13_99910339_99910626 | 0.18 |
GPR18 |
G protein-coupled receptor 18 |
146 |
0.96 |
| chr2_2650168_2650319 | 0.17 |
MYT1L |
myelin transcription factor 1-like |
315277 |
0.01 |
| chrY_15815919_15816489 | 0.17 |
TMSB4Y |
thymosin beta 4, Y-linked |
757 |
0.72 |
| chr13_84126063_84126363 | 0.17 |
ENSG00000222791 |
. |
253911 |
0.02 |
| chr13_41236803_41237000 | 0.17 |
FOXO1 |
forkhead box O1 |
3833 |
0.29 |
| chr1_197730742_197731002 | 0.16 |
RP11-448G4.4 |
|
4423 |
0.25 |
| chr4_123820303_123820454 | 0.16 |
NUDT6 |
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
14065 |
0.17 |
| chr13_31309966_31310295 | 0.16 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
485 |
0.88 |
| chr8_27131821_27132033 | 0.16 |
STMN4 |
stathmin-like 4 |
15990 |
0.18 |
| chr21_35306376_35306527 | 0.16 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
2933 |
0.21 |
| chr12_82826657_82826808 | 0.16 |
METTL25 |
methyltransferase like 25 |
33594 |
0.21 |
| chr7_129620661_129620812 | 0.16 |
ENSG00000263557 |
. |
4851 |
0.14 |
| chr1_162205339_162205490 | 0.16 |
ENSG00000266144 |
. |
78517 |
0.08 |
| chr15_96817385_96817602 | 0.15 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
1732 |
0.37 |
| chr5_118653769_118653920 | 0.15 |
ENSG00000243333 |
. |
11518 |
0.19 |
| chr1_114413652_114413958 | 0.15 |
PTPN22 |
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
515 |
0.69 |
| chr5_94384750_94385033 | 0.15 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
31700 |
0.22 |
| chr1_1099465_1099616 | 0.15 |
ENSG00000207730 |
. |
2944 |
0.11 |
| chr6_159464766_159465080 | 0.15 |
TAGAP |
T-cell activation RhoGTPase activating protein |
1127 |
0.51 |
| chr16_69598825_69599558 | 0.15 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
194 |
0.82 |
| chr1_246488003_246488154 | 0.15 |
SMYD3-IT1 |
SMYD3 intronic transcript 1 (non-protein coding) |
2338 |
0.35 |
| chr10_3514525_3515428 | 0.14 |
RP11-184A2.3 |
|
278283 |
0.01 |
| chr4_143488982_143489737 | 0.14 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
7537 |
0.34 |
| chr1_186292481_186292673 | 0.14 |
ENSG00000202025 |
. |
11517 |
0.17 |
| chr13_41555714_41555943 | 0.14 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
590 |
0.77 |
| chr1_198627689_198627922 | 0.14 |
RP11-553K8.5 |
|
8385 |
0.25 |
| chrX_64889806_64890147 | 0.14 |
MSN |
moesin |
2439 |
0.43 |
| chr20_43608188_43608641 | 0.14 |
STK4 |
serine/threonine kinase 4 |
13247 |
0.14 |
| chr2_64441014_64441273 | 0.14 |
AC074289.1 |
|
7034 |
0.28 |
| chr11_104904746_104905919 | 0.14 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
508 |
0.77 |
| chr3_189510811_189510962 | 0.14 |
TP63 |
tumor protein p63 |
3296 |
0.33 |
| chr1_198615140_198615291 | 0.14 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
6923 |
0.25 |
| chr21_36772703_36772854 | 0.14 |
ENSG00000211590 |
. |
320235 |
0.01 |
| chrX_54948821_54948972 | 0.14 |
TRO |
trophinin |
486 |
0.79 |
| chr9_137347658_137347902 | 0.14 |
RXRA |
retinoid X receptor, alpha |
49352 |
0.15 |
| chrX_25020829_25021251 | 0.14 |
ARX |
aristaless related homeobox |
13025 |
0.26 |
| chr3_13919763_13919914 | 0.13 |
WNT7A |
wingless-type MMTV integration site family, member 7A |
1780 |
0.41 |
| chr22_30040936_30041087 | 0.13 |
NF2 |
neurofibromin 2 (merlin) |
41023 |
0.11 |
| chrX_20996388_20996539 | 0.13 |
ENSG00000206716 |
. |
236292 |
0.02 |
| chr5_56496018_56496179 | 0.13 |
GPBP1 |
GC-rich promoter binding protein 1 |
13850 |
0.23 |
| chr15_60881660_60881911 | 0.13 |
RORA |
RAR-related orphan receptor A |
2955 |
0.3 |
| chr4_90226799_90226950 | 0.13 |
GPRIN3 |
GPRIN family member 3 |
2287 |
0.45 |
| chr6_37410160_37410312 | 0.13 |
CMTR1 |
cap methyltransferase 1 |
9240 |
0.22 |
| chr13_99910052_99910203 | 0.13 |
GPR18 |
G protein-coupled receptor 18 |
501 |
0.81 |
| chr6_30797258_30797409 | 0.13 |
ENSG00000202241 |
. |
34694 |
0.07 |
| chr6_82470488_82470639 | 0.13 |
ENSG00000206886 |
. |
3178 |
0.26 |
| chr1_169679248_169679951 | 0.13 |
SELL |
selectin L |
1240 |
0.48 |
| chr1_18010263_18010414 | 0.13 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
65489 |
0.12 |
| chr14_100535454_100535614 | 0.13 |
EVL |
Enah/Vasp-like |
2760 |
0.21 |
| chr14_22947593_22947744 | 0.12 |
TRAJ60 |
T cell receptor alpha joining 60 (pseudogene) |
2372 |
0.15 |
| chr15_81597008_81597159 | 0.12 |
IL16 |
interleukin 16 |
5326 |
0.21 |
| chr4_38623548_38623699 | 0.12 |
RP11-617D20.1 |
|
2573 |
0.31 |
| chr9_128586563_128586792 | 0.12 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
40873 |
0.21 |
| chr3_46411886_46412037 | 0.12 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
328 |
0.87 |
| chrY_17633910_17634061 | 0.12 |
ENSG00000252664 |
. |
540661 |
0.0 |
| chr11_119572371_119572522 | 0.12 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
26817 |
0.16 |
| chr12_9912992_9913534 | 0.12 |
CD69 |
CD69 molecule |
234 |
0.92 |
| chr20_35575234_35575857 | 0.12 |
SAMHD1 |
SAM domain and HD domain 1 |
4566 |
0.25 |
| chr7_140014228_140014507 | 0.12 |
SLC37A3 |
solute carrier family 37, member 3 |
28922 |
0.14 |
| chr1_185284312_185284463 | 0.12 |
IVNS1ABP |
influenza virus NS1A binding protein |
2074 |
0.35 |
| chr7_26141262_26141558 | 0.12 |
ENSG00000266430 |
. |
41451 |
0.15 |
| chr17_2286013_2286164 | 0.12 |
RP1-59D14.1 |
|
2053 |
0.18 |
| chr19_42390602_42391041 | 0.12 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
2306 |
0.19 |
| chr11_42259823_42259974 | 0.12 |
LRRC4C |
leucine rich repeat containing 4C |
778575 |
0.0 |
| chr4_170256258_170256409 | 0.12 |
SH3RF1 |
SH3 domain containing ring finger 1 |
64077 |
0.14 |
| chr2_194212623_194212914 | 0.12 |
NA |
NA |
> 106 |
NA |
| chr13_38923188_38923622 | 0.12 |
UFM1 |
ubiquitin-fold modifier 1 |
583 |
0.86 |
| chr17_264289_264440 | 0.12 |
AC108004.3 |
|
550 |
0.69 |
| chr6_154568354_154568864 | 0.12 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
53 |
0.99 |
| chr15_94040362_94040513 | 0.12 |
ENSG00000212063 |
. |
210885 |
0.02 |
| chr5_81074467_81075538 | 0.11 |
SSBP2 |
single-stranded DNA binding protein 2 |
27930 |
0.25 |
| chr2_28826434_28826585 | 0.11 |
PLB1 |
phospholipase B1 |
1723 |
0.42 |
| chr17_38018344_38018495 | 0.11 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
1960 |
0.26 |
| chr10_123458202_123458353 | 0.11 |
RP11-78A18.2 |
|
37541 |
0.2 |
| chr3_32998333_32998484 | 0.11 |
CCR4 |
chemokine (C-C motif) receptor 4 |
5342 |
0.28 |
| chr21_32559881_32560032 | 0.11 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
57417 |
0.15 |
| chrX_803799_804090 | 0.11 |
SHOX |
short stature homeobox |
212402 |
0.02 |
| chrY_753799_754090 | 0.11 |
NA |
NA |
> 106 |
NA |
| chr5_106791734_106791885 | 0.11 |
EFNA5 |
ephrin-A5 |
214519 |
0.02 |
| chr1_200865204_200865355 | 0.11 |
C1orf106 |
chromosome 1 open reading frame 106 |
1330 |
0.42 |
| chr6_26198173_26198564 | 0.11 |
HIST1H3D |
histone cluster 1, H3d |
890 |
0.22 |
| chr2_37576303_37576454 | 0.11 |
QPCT |
glutaminyl-peptide cyclotransferase |
4526 |
0.2 |
| chr9_102849500_102849651 | 0.11 |
ERP44 |
endoplasmic reticulum protein 44 |
11747 |
0.18 |
| chr4_61665381_61665532 | 0.11 |
ENSG00000265829 |
. |
122881 |
0.06 |
| chr14_99726303_99726510 | 0.11 |
AL109767.1 |
|
2879 |
0.29 |
| chr2_197032053_197032740 | 0.11 |
STK17B |
serine/threonine kinase 17b |
3328 |
0.25 |
| chr1_24862040_24862255 | 0.11 |
ENSG00000266551 |
. |
5943 |
0.18 |
| chr20_58406416_58406567 | 0.11 |
ENSG00000238777 |
. |
21292 |
0.2 |
| chr18_76745276_76745427 | 0.11 |
SALL3 |
spalt-like transcription factor 3 |
5076 |
0.32 |
| chr3_20050398_20050549 | 0.11 |
PP2D1 |
protein phosphatase 2C-like domain containing 1 |
3349 |
0.22 |
| chr6_106972751_106973424 | 0.11 |
AIM1 |
absent in melanoma 1 |
13357 |
0.2 |
| chr6_17703935_17704086 | 0.11 |
RP11-500C11.3 |
|
2478 |
0.22 |
| chr9_20242645_20242902 | 0.11 |
ENSG00000221744 |
. |
52229 |
0.16 |
| chr18_3329157_3329308 | 0.11 |
MYL12B |
myosin, light chain 12B, regulatory |
66278 |
0.09 |
| chrY_2804248_2804778 | 0.11 |
ZFY |
zinc finger protein, Y-linked |
967 |
0.67 |
| chr17_47819247_47819793 | 0.11 |
FAM117A |
family with sequence similarity 117, member A |
17631 |
0.14 |
| chr17_40700695_40700889 | 0.11 |
HSD17B1 |
hydroxysteroid (17-beta) dehydrogenase 1 |
440 |
0.63 |
| chr1_25446753_25447069 | 0.11 |
RP4-706G24.1 |
|
87719 |
0.07 |
| chr6_138192975_138193775 | 0.11 |
RP11-356I2.4 |
|
4005 |
0.25 |
| chr16_18957307_18957806 | 0.11 |
ENSG00000265515 |
. |
15952 |
0.13 |
| chr17_75456326_75456913 | 0.11 |
SEPT9 |
septin 9 |
4171 |
0.18 |
| chr1_43151006_43151188 | 0.11 |
YBX1 |
Y box binding protein 1 |
2439 |
0.25 |
| chr3_151961363_151961565 | 0.11 |
MBNL1 |
muscleblind-like splicing regulator 1 |
24365 |
0.2 |
| chr8_113941673_113941824 | 0.11 |
CSMD3 |
CUB and Sushi multiple domains 3 |
239477 |
0.02 |
| chr14_99708318_99708739 | 0.11 |
AL109767.1 |
|
20757 |
0.2 |
| chr2_143916963_143917114 | 0.10 |
RP11-190J23.1 |
|
12703 |
0.25 |
| chr17_76335263_76335414 | 0.10 |
SOCS3 |
suppressor of cytokine signaling 3 |
20817 |
0.14 |
| chr1_16483683_16483834 | 0.10 |
EPHA2 |
EPH receptor A2 |
1176 |
0.29 |
| chr12_27974048_27974199 | 0.10 |
ENSG00000201612 |
. |
14800 |
0.14 |
| chr4_41188158_41188349 | 0.10 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
28222 |
0.17 |
| chr3_171848594_171848745 | 0.10 |
FNDC3B |
fibronectin type III domain containing 3B |
3905 |
0.33 |
| chr15_64767992_64768143 | 0.10 |
RP11-702L15.4 |
|
5615 |
0.15 |
| chr5_159900576_159900727 | 0.10 |
ENSG00000265237 |
. |
758 |
0.62 |
| chr13_48988954_48989249 | 0.10 |
LPAR6 |
lysophosphatidic acid receptor 6 |
11942 |
0.27 |
| chr3_32996346_32996497 | 0.10 |
CCR4 |
chemokine (C-C motif) receptor 4 |
3355 |
0.33 |
| chr3_114010788_114010939 | 0.10 |
TIGIT |
T cell immunoreceptor with Ig and ITIM domains |
914 |
0.57 |
| chr21_32532372_32532523 | 0.10 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
29908 |
0.23 |
| chr7_106507057_106507208 | 0.10 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
1208 |
0.59 |
| chr4_124343109_124343622 | 0.10 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
22242 |
0.29 |
| chr15_30213010_30213162 | 0.10 |
TJP1 |
tight junction protein 1 |
47982 |
0.15 |
| chr3_71111068_71111292 | 0.10 |
FOXP1 |
forkhead box P1 |
2897 |
0.41 |
| chr4_95378812_95378990 | 0.10 |
PDLIM5 |
PDZ and LIM domain 5 |
2505 |
0.43 |
| chr15_34634999_34635332 | 0.10 |
NOP10 |
NOP10 ribonucleoprotein |
159 |
0.71 |
| chr1_244504765_244505074 | 0.10 |
C1orf100 |
chromosome 1 open reading frame 100 |
11018 |
0.25 |
| chr12_65779210_65779361 | 0.10 |
MSRB3 |
methionine sulfoxide reductase B3 |
58630 |
0.15 |
| chr10_30745504_30745655 | 0.10 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
17828 |
0.21 |
| chr3_108543496_108543820 | 0.10 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
2039 |
0.42 |
| chr5_85275732_85275883 | 0.10 |
NBPF22P |
neuroblastoma breakpoint family, member 22, pseudogene |
302777 |
0.01 |
| chr10_129861850_129862130 | 0.10 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
16156 |
0.26 |
| chr22_31615175_31615326 | 0.10 |
ENSG00000199695 |
. |
3589 |
0.13 |
| chr14_26016058_26016209 | 0.10 |
ENSG00000212270 |
. |
259785 |
0.02 |
| chr1_150547532_150547683 | 0.10 |
MCL1 |
myeloid cell leukemia sequence 1 (BCL2-related) |
4399 |
0.09 |
| chr8_112438072_112438223 | 0.10 |
ENSG00000222146 |
. |
722907 |
0.0 |
| chr4_129450192_129450343 | 0.10 |
ENSG00000238802 |
. |
226914 |
0.02 |
| chr17_33374319_33374470 | 0.10 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
5024 |
0.13 |
| chr15_52361483_52361634 | 0.10 |
CTD-2184D3.5 |
|
31162 |
0.12 |
| chr13_41227499_41227650 | 0.10 |
FOXO1 |
forkhead box O1 |
13160 |
0.24 |
| chr21_32555155_32555306 | 0.10 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
52691 |
0.16 |
| chr14_22966536_22967037 | 0.10 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
10615 |
0.1 |
| chr12_53773389_53773606 | 0.10 |
SP1 |
Sp1 transcription factor |
463 |
0.73 |
| chr18_67822225_67822376 | 0.10 |
RTTN |
rotatin |
50662 |
0.17 |
| chr6_44899717_44900076 | 0.10 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
23351 |
0.28 |
| chr5_94951954_94952105 | 0.10 |
GPR150 |
G protein-coupled receptor 150 |
3753 |
0.21 |
| chr4_140587624_140588180 | 0.10 |
MGST2 |
microsomal glutathione S-transferase 2 |
913 |
0.61 |
| chr9_97864832_97864983 | 0.10 |
RP11-80I15.4 |
|
11083 |
0.14 |
| chr2_192623983_192624134 | 0.10 |
AC098872.3 |
|
34093 |
0.19 |
| chr15_101783225_101783409 | 0.09 |
CHSY1 |
chondroitin sulfate synthase 1 |
8820 |
0.18 |
| chr12_103834650_103834908 | 0.09 |
C12orf42 |
chromosome 12 open reading frame 42 |
54952 |
0.15 |
| chr1_100852790_100852941 | 0.09 |
ENSG00000216067 |
. |
8534 |
0.21 |
| chr4_47491811_47491962 | 0.09 |
ATP10D |
ATPase, class V, type 10D |
4570 |
0.24 |
| chrX_31453419_31453570 | 0.09 |
ENSG00000252903 |
. |
87185 |
0.1 |
| chr2_68961577_68961872 | 0.09 |
ARHGAP25 |
Rho GTPase activating protein 25 |
189 |
0.96 |
| chr14_22182172_22182323 | 0.09 |
OR4E2 |
olfactory receptor, family 4, subfamily E, member 2 |
48950 |
0.13 |
| chr7_32929941_32930760 | 0.09 |
KBTBD2 |
kelch repeat and BTB (POZ) domain containing 2 |
415 |
0.87 |
| chr7_150217655_150217806 | 0.09 |
GIMAP7 |
GTPase, IMAP family member 7 |
5812 |
0.21 |
| chr3_151963607_151963763 | 0.09 |
MBNL1 |
muscleblind-like splicing regulator 1 |
22144 |
0.21 |
| chr5_36951473_36951624 | 0.09 |
NIPBL |
Nipped-B homolog (Drosophila) |
74656 |
0.12 |
| chr10_64146533_64146728 | 0.09 |
ZNF365 |
zinc finger protein 365 |
11327 |
0.25 |
| chr12_47604114_47604265 | 0.09 |
PCED1B |
PC-esterase domain containing 1B |
5863 |
0.23 |
| chr8_65492553_65493166 | 0.09 |
BHLHE22 |
basic helix-loop-helix family, member e22 |
45 |
0.97 |
| chr11_64089315_64089466 | 0.09 |
PRDX5 |
peroxiredoxin 5 |
3736 |
0.08 |
| chr16_48642312_48642839 | 0.09 |
N4BP1 |
NEDD4 binding protein 1 |
1545 |
0.4 |
| chr6_22436672_22436823 | 0.09 |
HDGFL1 |
hepatoma derived growth factor-like 1 |
132931 |
0.05 |
| chr1_186288985_186289136 | 0.09 |
ENSG00000202025 |
. |
8000 |
0.18 |
| chr2_192176773_192176924 | 0.09 |
MYO1B |
myosin IB |
35237 |
0.19 |
| chr18_3453699_3454909 | 0.09 |
TGIF1 |
TGFB-induced factor homeobox 1 |
532 |
0.8 |
| chr9_41432727_41433091 | 0.09 |
SPATA31A5 |
SPATA31 subfamily A, member 5 |
67770 |
0.13 |
| chr10_63999686_63999837 | 0.09 |
RTKN2 |
rhotekin 2 |
3739 |
0.34 |
| chr7_43688967_43689164 | 0.09 |
COA1 |
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
830 |
0.67 |
| chr4_39032194_39032374 | 0.09 |
TMEM156 |
transmembrane protein 156 |
1757 |
0.38 |
| chr17_78755045_78755196 | 0.09 |
RP11-28G8.1 |
|
24312 |
0.21 |
| chr2_174129320_174129481 | 0.09 |
MLK7-AS1 |
MLK7 antisense RNA 1 |
6948 |
0.3 |
| chr5_44684027_44684178 | 0.09 |
ENSG00000263556 |
. |
32190 |
0.23 |
| chr4_176699040_176699191 | 0.09 |
GPM6A |
glycoprotein M6A |
9423 |
0.24 |
| chr19_42056193_42056486 | 0.09 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
453 |
0.8 |
| chr16_75182070_75182247 | 0.09 |
ZFP1 |
ZFP1 zinc finger protein |
232 |
0.91 |
| chr6_10522420_10522571 | 0.09 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
911 |
0.58 |
| chr11_47535258_47535409 | 0.09 |
RP11-750H9.7 |
|
1108 |
0.33 |
| chrX_124911556_124911707 | 0.09 |
ENSG00000222518 |
. |
114732 |
0.07 |
| chr9_100691376_100691527 | 0.09 |
C9orf156 |
chromosome 9 open reading frame 156 |
6599 |
0.16 |
| chr10_91151882_91152232 | 0.09 |
IFIT1 |
interferon-induced protein with tetratricopeptide repeats 1 |
246 |
0.89 |
| chr2_85515182_85515333 | 0.09 |
ENSG00000221579 |
. |
10900 |
0.13 |
| chr2_61111507_61111691 | 0.09 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
2808 |
0.27 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.1 | 0.2 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.3 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0060044 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0043921 | modulation by host of viral transcription(GO:0043921) modulation by host of symbiont transcription(GO:0052472) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0044409 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.0 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.8 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |