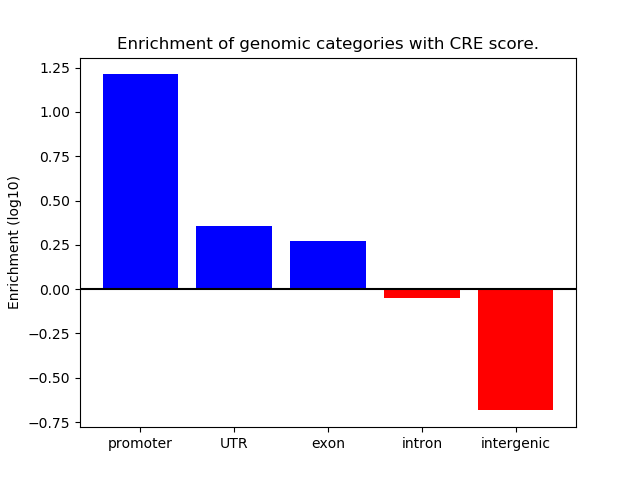
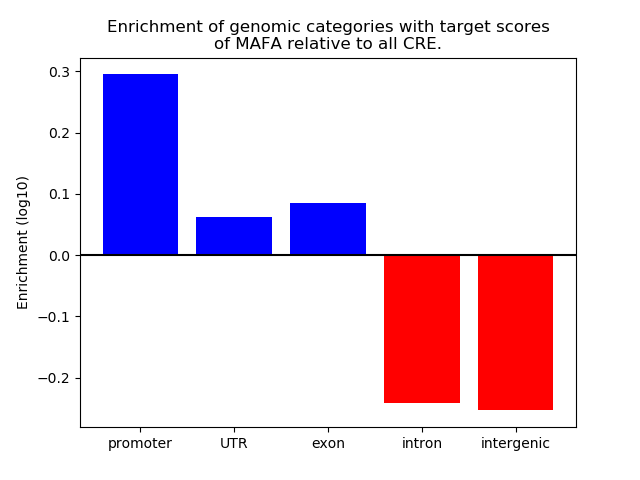
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
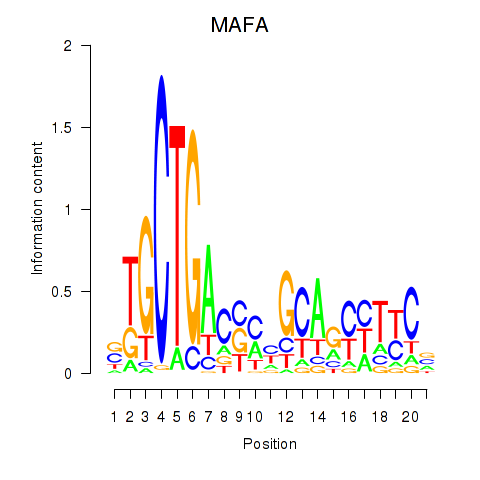
Results for MAFA
Z-value: 1.01
Transcription factors associated with MAFA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFA
|
ENSG00000182759.3 | MAF bZIP transcription factor A |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_144510266_144510417 | MAFA | 2235 | 0.183325 | 0.77 | 1.4e-02 | Click! |
| chr8_144543935_144544113 | MAFA | 31448 | 0.084746 | 0.76 | 1.8e-02 | Click! |
| chr8_144544411_144544712 | MAFA | 31985 | 0.083952 | 0.72 | 3.0e-02 | Click! |
| chr8_144510497_144510807 | MAFA | 1924 | 0.207447 | 0.71 | 3.4e-02 | Click! |
| chr8_144544755_144544972 | MAFA | 32287 | 0.083504 | 0.71 | 3.4e-02 | Click! |
Activity of the MAFA motif across conditions
Conditions sorted by the z-value of the MAFA motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_40684769_40685057 | 0.56 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
5313 |
0.23 |
| chr14_100533458_100533776 | 0.53 |
EVL |
Enah/Vasp-like |
843 |
0.55 |
| chr16_27414444_27414647 | 0.48 |
IL21R |
interleukin 21 receptor |
122 |
0.97 |
| chrY_7207933_7208104 | 0.46 |
ENSG00000251996 |
. |
1554 |
0.4 |
| chr7_150148386_150148563 | 0.46 |
GIMAP8 |
GTPase, IMAP family member 8 |
756 |
0.63 |
| chr10_6621928_6622130 | 0.39 |
PRKCQ |
protein kinase C, theta |
172 |
0.98 |
| chr14_69095479_69095684 | 0.39 |
CTD-2325P2.4 |
|
419 |
0.89 |
| chr16_87799045_87799225 | 0.39 |
KLHDC4 |
kelch domain containing 4 |
370 |
0.86 |
| chr16_3628507_3629057 | 0.39 |
NLRC3 |
NLR family, CARD domain containing 3 |
1381 |
0.35 |
| chr3_81545471_81545622 | 0.38 |
ENSG00000222389 |
. |
13081 |
0.32 |
| chr13_99222825_99223522 | 0.37 |
STK24 |
serine/threonine kinase 24 |
5944 |
0.22 |
| chr16_89788020_89788196 | 0.36 |
ZNF276 |
zinc finger protein 276 |
156 |
0.84 |
| chr11_16946573_16946796 | 0.35 |
PLEKHA7 |
pleckstrin homology domain containing, family A member 7 |
43779 |
0.12 |
| chr1_145440326_145440711 | 0.35 |
TXNIP |
thioredoxin interacting protein |
1212 |
0.33 |
| chr19_6501621_6501878 | 0.34 |
TUBB4A |
tubulin, beta 4A class IVa |
49 |
0.95 |
| chrX_123481094_123481375 | 0.34 |
SH2D1A |
SH2 domain containing 1A |
795 |
0.78 |
| chr17_76123138_76123289 | 0.34 |
TMC6 |
transmembrane channel-like 6 |
112 |
0.94 |
| chr8_134080772_134080923 | 0.34 |
SLA |
Src-like-adaptor |
8244 |
0.25 |
| chr19_49931722_49931873 | 0.33 |
GFY |
golgi-associated, olfactory signaling regulator |
4791 |
0.07 |
| chr1_36272671_36272822 | 0.32 |
AGO4 |
argonaute RISC catalytic component 4 |
1027 |
0.53 |
| chr9_503761_503951 | 0.32 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
860 |
0.51 |
| chr20_37435342_37435890 | 0.32 |
PPP1R16B |
protein phosphatase 1, regulatory subunit 16B |
1249 |
0.47 |
| chr21_46346899_46347050 | 0.32 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
1659 |
0.21 |
| chr11_63656035_63656583 | 0.32 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
141 |
0.94 |
| chr12_92172716_92172989 | 0.31 |
C12orf79 |
chromosome 12 open reading frame 79 |
357945 |
0.01 |
| chr7_3083968_3084119 | 0.31 |
CARD11 |
caspase recruitment domain family, member 11 |
464 |
0.86 |
| chr20_61148226_61148454 | 0.30 |
C20orf166-AS1 |
C20orf166 antisense RNA 1 |
378 |
0.63 |
| chr19_41260606_41261007 | 0.30 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
3692 |
0.12 |
| chr4_40201867_40202018 | 0.29 |
RHOH |
ras homolog family member H |
22 |
0.98 |
| chr19_37808857_37809211 | 0.29 |
HKR1 |
HKR1, GLI-Kruppel zinc finger family member |
93 |
0.97 |
| chr12_54891563_54891796 | 0.29 |
NCKAP1L |
NCK-associated protein 1-like |
184 |
0.93 |
| chr17_76128459_76128696 | 0.29 |
TMC6 |
transmembrane channel-like 6 |
89 |
0.94 |
| chr9_100000727_100000985 | 0.29 |
CCDC180 |
coiled-coil domain containing 180 |
77 |
0.98 |
| chr1_15249911_15250062 | 0.29 |
KAZN |
kazrin, periplakin interacting protein |
640 |
0.83 |
| chr3_71478030_71478538 | 0.29 |
ENSG00000221264 |
. |
112956 |
0.06 |
| chr3_196365832_196366436 | 0.29 |
NRROS |
negative regulator of reactive oxygen species |
423 |
0.51 |
| chr10_8103703_8104121 | 0.28 |
GATA3 |
GATA binding protein 3 |
7143 |
0.33 |
| chr13_41187779_41187930 | 0.28 |
FOXO1 |
forkhead box O1 |
52880 |
0.14 |
| chr13_41495302_41495814 | 0.28 |
SUGT1P3 |
SUGT1 pseudogene 3 |
289 |
0.53 |
| chr2_149634276_149634517 | 0.28 |
KIF5C |
kinesin family member 5C |
1577 |
0.38 |
| chr6_89790739_89791095 | 0.27 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
420 |
0.52 |
| chrX_39185008_39185159 | 0.27 |
ENSG00000207122 |
. |
244545 |
0.02 |
| chr20_62271176_62271486 | 0.27 |
CTD-3184A7.4 |
|
12728 |
0.09 |
| chr5_35797219_35797401 | 0.27 |
SPEF2 |
sperm flagellar 2 |
18040 |
0.19 |
| chr19_54880943_54881230 | 0.27 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
972 |
0.39 |
| chr19_3179462_3180177 | 0.27 |
S1PR4 |
sphingosine-1-phosphate receptor 4 |
1083 |
0.38 |
| chr14_22631179_22631341 | 0.27 |
ENSG00000238634 |
. |
20373 |
0.25 |
| chr1_111743477_111743628 | 0.26 |
CHI3L2 |
chitinase 3-like 2 |
159 |
0.6 |
| chr3_18466629_18466793 | 0.26 |
SATB1 |
SATB homeobox 1 |
118 |
0.97 |
| chr1_155939445_155939596 | 0.26 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
0 |
0.95 |
| chr18_33196454_33196850 | 0.26 |
RP11-712P20.2 |
|
20787 |
0.21 |
| chrX_65294918_65295209 | 0.26 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
35096 |
0.18 |
| chr12_65064432_65064583 | 0.26 |
RP11-338E21.3 |
|
15708 |
0.13 |
| chr12_6570416_6570672 | 0.26 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
9267 |
0.08 |
| chr13_24826013_24826241 | 0.25 |
SPATA13 |
spermatogenesis associated 13 |
271 |
0.9 |
| chr1_211503167_211503447 | 0.25 |
TRAF5 |
TNF receptor-associated factor 5 |
3128 |
0.31 |
| chr4_90228457_90229530 | 0.25 |
GPRIN3 |
GPRIN family member 3 |
168 |
0.97 |
| chr2_203499531_203499682 | 0.25 |
FAM117B |
family with sequence similarity 117, member B |
295 |
0.94 |
| chr12_54812737_54812923 | 0.25 |
ITGA5 |
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
414 |
0.6 |
| chr19_51228076_51228227 | 0.25 |
CLEC11A |
C-type lectin domain family 11, member A |
1371 |
0.27 |
| chr22_33278140_33278291 | 0.25 |
TIMP3 |
TIMP metallopeptidase inhibitor 3 |
80528 |
0.1 |
| chr17_54911768_54911997 | 0.25 |
DGKE |
diacylglycerol kinase, epsilon 64kDa |
414 |
0.59 |
| chr10_135170371_135170656 | 0.25 |
FUOM |
fucose mutarotase |
728 |
0.41 |
| chr22_24038685_24038889 | 0.25 |
RGL4 |
ral guanine nucleotide dissociation stimulator-like 4 |
116 |
0.94 |
| chr17_72755086_72755237 | 0.25 |
SLC9A3R1 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
2981 |
0.14 |
| chr9_131550054_131550269 | 0.25 |
TBC1D13 |
TBC1 domain family, member 13 |
533 |
0.64 |
| chr1_9686934_9687157 | 0.24 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
24745 |
0.14 |
| chr1_110045467_110045747 | 0.24 |
AMIGO1 |
adhesion molecule with Ig-like domain 1 |
6697 |
0.11 |
| chr1_12238656_12238807 | 0.24 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
11671 |
0.15 |
| chr13_103426260_103426509 | 0.24 |
TEX30 |
testis expressed 30 |
223 |
0.89 |
| chr2_65356875_65357219 | 0.24 |
RAB1A |
RAB1A, member RAS oncogene family |
188 |
0.95 |
| chr20_3996304_3996483 | 0.24 |
RNF24 |
ring finger protein 24 |
164 |
0.96 |
| chr12_22487067_22487257 | 0.24 |
ST8SIA1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
31 |
0.99 |
| chr15_45008032_45008258 | 0.24 |
B2M |
beta-2-microglobulin |
4430 |
0.17 |
| chr4_169799366_169799588 | 0.24 |
ENSG00000201176 |
. |
7029 |
0.22 |
| chr5_139493535_139493731 | 0.23 |
PURA |
purine-rich element binding protein A |
75 |
0.96 |
| chr7_1978940_1979381 | 0.23 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
1025 |
0.64 |
| chr13_99992644_99992897 | 0.23 |
ENSG00000207719 |
. |
15615 |
0.19 |
| chr2_43354715_43354866 | 0.23 |
ENSG00000207087 |
. |
36158 |
0.2 |
| chr10_135089035_135089208 | 0.23 |
ADAM8 |
ADAM metallopeptidase domain 8 |
1233 |
0.29 |
| chr20_35575234_35575857 | 0.23 |
SAMHD1 |
SAM domain and HD domain 1 |
4566 |
0.25 |
| chr8_94930047_94930344 | 0.23 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
220 |
0.92 |
| chr19_17447799_17448161 | 0.23 |
GTPBP3 |
GTP binding protein 3 (mitochondrial) |
373 |
0.68 |
| chrX_153190817_153191535 | 0.23 |
ARHGAP4 |
Rho GTPase activating protein 4 |
522 |
0.6 |
| chr2_204191257_204191408 | 0.23 |
ABI2 |
abl-interactor 2 |
1610 |
0.37 |
| chr19_17961513_17961726 | 0.23 |
JAK3 |
Janus kinase 3 |
2778 |
0.16 |
| chr8_24799151_24799367 | 0.23 |
CTD-2168K21.2 |
|
14876 |
0.17 |
| chr2_136873940_136874440 | 0.22 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
377 |
0.92 |
| chr9_134139798_134140094 | 0.22 |
FAM78A |
family with sequence similarity 78, member A |
5934 |
0.19 |
| chr1_6613959_6614285 | 0.22 |
NOL9 |
nucleolar protein 9 |
473 |
0.62 |
| chr6_20402144_20402837 | 0.22 |
E2F3 |
E2F transcription factor 3 |
92 |
0.96 |
| chr22_21823868_21824019 | 0.22 |
PI4KAP2 |
phosphatidylinositol 4-kinase, catalytic, alpha pseudogene 2 |
18706 |
0.11 |
| chr20_43608188_43608641 | 0.22 |
STK4 |
serine/threonine kinase 4 |
13247 |
0.14 |
| chrX_129225814_129226276 | 0.22 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
18291 |
0.19 |
| chr21_36901015_36901338 | 0.22 |
ENSG00000211590 |
. |
191837 |
0.03 |
| chr1_27847952_27848103 | 0.22 |
RP1-159A19.4 |
|
4289 |
0.2 |
| chr15_26104297_26104448 | 0.22 |
ATP10A |
ATPase, class V, type 10A |
3983 |
0.23 |
| chr7_2561879_2562201 | 0.22 |
LFNG |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
2544 |
0.22 |
| chr20_47411194_47411345 | 0.22 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
33151 |
0.2 |
| chrX_107070045_107070650 | 0.22 |
MID2 |
midline 2 |
703 |
0.7 |
| chr6_135518404_135518725 | 0.21 |
MYB-AS1 |
MYB antisense RNA 1 |
1431 |
0.4 |
| chr7_153749645_153749898 | 0.21 |
DPP6 |
dipeptidyl-peptidase 6 |
6 |
0.98 |
| chr13_100072782_100072933 | 0.21 |
ENSG00000266207 |
. |
32866 |
0.16 |
| chr17_41712002_41712153 | 0.21 |
MEOX1 |
mesenchyme homeobox 1 |
26854 |
0.14 |
| chr13_23754987_23755138 | 0.21 |
SGCG |
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
29 |
0.98 |
| chr17_38720511_38720770 | 0.21 |
CCR7 |
chemokine (C-C motif) receptor 7 |
1071 |
0.49 |
| chr5_130617597_130618118 | 0.21 |
CDC42SE2 |
CDC42 small effector 2 |
18064 |
0.27 |
| chr6_143178538_143178701 | 0.21 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
20435 |
0.26 |
| chr6_150184570_150184721 | 0.21 |
RP11-244K5.8 |
|
15 |
0.89 |
| chrX_55478600_55478841 | 0.21 |
MAGEH1 |
melanoma antigen family H, 1 |
182 |
0.88 |
| chr17_45812120_45812293 | 0.21 |
TBX21 |
T-box 21 |
1596 |
0.32 |
| chr5_172111392_172111616 | 0.21 |
CTB-79E8.2 |
|
13219 |
0.15 |
| chr2_5727897_5728048 | 0.21 |
AC108025.2 |
|
103206 |
0.07 |
| chr10_73847731_73848399 | 0.21 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
21 |
0.98 |
| chr22_21058522_21058673 | 0.20 |
POM121L4P |
POM121 transmembrane nucleoporin-like 4 pseudogene |
14380 |
0.13 |
| chr11_67173386_67173537 | 0.20 |
TBC1D10C |
TBC1 domain family, member 10C |
1801 |
0.14 |
| chr9_132565902_132566404 | 0.20 |
TOR1B |
torsin family 1, member B (torsin B) |
606 |
0.67 |
| chr6_159331017_159331168 | 0.20 |
ENSG00000223191 |
. |
16511 |
0.18 |
| chr1_161038546_161039545 | 0.20 |
ARHGAP30 |
Rho GTPase activating protein 30 |
411 |
0.65 |
| chr10_53458598_53459393 | 0.20 |
CSTF2T |
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
360 |
0.92 |
| chr10_123524520_123524671 | 0.20 |
RP11-78A18.2 |
|
28777 |
0.22 |
| chr16_89427901_89428186 | 0.20 |
ANKRD11 |
ankyrin repeat domain 11 |
32649 |
0.1 |
| chr6_37400508_37400927 | 0.20 |
CMTR1 |
cap methyltransferase 1 |
279 |
0.92 |
| chrX_134232377_134232653 | 0.20 |
FAM127B |
family with sequence similarity 127, member B |
46310 |
0.12 |
| chr12_113622602_113622792 | 0.20 |
DDX54 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
587 |
0.4 |
| chr10_102588210_102588406 | 0.20 |
ENSG00000222072 |
. |
78729 |
0.07 |
| chr1_214454566_214454774 | 0.20 |
SMYD2 |
SET and MYND domain containing 2 |
94 |
0.98 |
| chr12_21654786_21654996 | 0.20 |
GOLT1B |
golgi transport 1B |
142 |
0.69 |
| chr3_47029106_47029289 | 0.20 |
CCDC12 |
coiled-coil domain containing 12 |
5725 |
0.19 |
| chr20_10153323_10153603 | 0.20 |
SNAP25 |
synaptosomal-associated protein, 25kDa |
46015 |
0.15 |
| chr19_2553269_2553420 | 0.20 |
ENSG00000252962 |
. |
50205 |
0.09 |
| chr1_211501150_211501529 | 0.20 |
TRAF5 |
TNF receptor-associated factor 5 |
1160 |
0.57 |
| chr4_148653403_148653650 | 0.20 |
ARHGAP10 |
Rho GTPase activating protein 10 |
312 |
0.89 |
| chr9_38424210_38424397 | 0.20 |
IGFBPL1 |
insulin-like growth factor binding protein-like 1 |
141 |
0.97 |
| chr22_19879923_19880332 | 0.20 |
GNB1L |
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
37665 |
0.1 |
| chr1_64059062_64059382 | 0.19 |
PGM1 |
phosphoglucomutase 1 |
258 |
0.92 |
| chr16_28998125_28998276 | 0.19 |
LAT |
linker for activation of T cells |
667 |
0.47 |
| chr17_75453210_75453403 | 0.19 |
SEPT9 |
septin 9 |
858 |
0.54 |
| chr9_125667319_125667538 | 0.19 |
RC3H2 |
ring finger and CCCH-type domains 2 |
66 |
0.95 |
| chr10_30818195_30818576 | 0.19 |
ENSG00000239744 |
. |
26448 |
0.21 |
| chr22_18255678_18255932 | 0.19 |
BID |
BH3 interacting domain death agonist |
977 |
0.55 |
| chr17_36715792_36715943 | 0.19 |
SRCIN1 |
SRC kinase signaling inhibitor 1 |
3793 |
0.19 |
| chr19_41833582_41833733 | 0.19 |
CCDC97 |
coiled-coil domain containing 97 |
8019 |
0.1 |
| chr8_132916838_132917136 | 0.19 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
631 |
0.84 |
| chr14_99655246_99655958 | 0.19 |
AL162151.4 |
|
30849 |
0.2 |
| chr17_38020283_38020945 | 0.19 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
173 |
0.93 |
| chr14_105955874_105956035 | 0.19 |
C14orf80 |
chromosome 14 open reading frame 80 |
238 |
0.86 |
| chr5_140983442_140983851 | 0.19 |
AC008781.7 |
|
14335 |
0.11 |
| chr20_3747782_3747941 | 0.19 |
C20orf27 |
chromosome 20 open reading frame 27 |
514 |
0.68 |
| chr6_159483760_159483976 | 0.19 |
TAGAP |
T-cell activation RhoGTPase activating protein |
17684 |
0.19 |
| chr5_43034344_43034652 | 0.19 |
AC025171.1 |
|
7179 |
0.15 |
| chr2_74214423_74214717 | 0.19 |
DGUOK-AS1 |
DGUOK antisense RNA 1 |
6002 |
0.17 |
| chr17_62207148_62207496 | 0.19 |
ERN1 |
endoplasmic reticulum to nucleus signaling 1 |
163 |
0.95 |
| chr2_87829980_87830131 | 0.19 |
RP11-1399P15.1 |
|
52502 |
0.16 |
| chr19_3136242_3137376 | 0.19 |
GNA15 |
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
618 |
0.58 |
| chr12_124873885_124874250 | 0.19 |
NCOR2 |
nuclear receptor corepressor 2 |
697 |
0.79 |
| chr11_118087227_118087423 | 0.19 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
1719 |
0.29 |
| chr7_50728203_50728447 | 0.19 |
GRB10 |
growth factor receptor-bound protein 10 |
33698 |
0.21 |
| chr2_127783302_127783743 | 0.19 |
ENSG00000238788 |
. |
8525 |
0.27 |
| chr20_39769733_39769905 | 0.19 |
RP1-1J6.2 |
|
3176 |
0.25 |
| chr19_3275516_3275954 | 0.18 |
AC010649.1 |
|
27184 |
0.14 |
| chr8_146127330_146127682 | 0.18 |
ZNF250 |
zinc finger protein 250 |
47 |
0.97 |
| chr14_78174355_78174506 | 0.18 |
SLIRP |
SRA stem-loop interacting RNA binding protein |
8 |
0.54 |
| chr3_45152218_45152388 | 0.18 |
CDCP1 |
CUB domain containing protein 1 |
35611 |
0.16 |
| chr5_134181747_134182027 | 0.18 |
C5orf24 |
chromosome 5 open reading frame 24 |
43 |
0.97 |
| chr1_233085968_233086196 | 0.18 |
NTPCR |
nucleoside-triphosphatase, cancer-related |
288 |
0.94 |
| chr8_86132724_86133084 | 0.18 |
C8orf59 |
chromosome 8 open reading frame 59 |
254 |
0.88 |
| chr18_60383353_60383643 | 0.18 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
815 |
0.66 |
| chr7_150180608_150181504 | 0.18 |
GIMAP7 |
GTPase, IMAP family member 7 |
30862 |
0.13 |
| chr16_71930385_71930591 | 0.18 |
IST1 |
increased sodium tolerance 1 homolog (yeast) |
996 |
0.42 |
| chr10_32667341_32667675 | 0.18 |
EPC1 |
enhancer of polycomb homolog 1 (Drosophila) |
218 |
0.91 |
| chr12_4377755_4378334 | 0.18 |
CCND2 |
cyclin D2 |
4894 |
0.19 |
| chr12_122230356_122230788 | 0.18 |
RHOF |
ras homolog family member F (in filopodia) |
694 |
0.64 |
| chr6_33244320_33244875 | 0.18 |
B3GALT4 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
320 |
0.73 |
| chr19_4370207_4370410 | 0.18 |
AC007292.6 |
|
6522 |
0.08 |
| chr17_39781410_39781561 | 0.18 |
KRT17 |
keratin 17 |
391 |
0.74 |
| chr8_145447065_145447230 | 0.18 |
FAM203B |
family with sequence similarity 203, member B |
8277 |
0.11 |
| chr1_6418667_6418818 | 0.18 |
ACOT7 |
acyl-CoA thioesterase 7 |
662 |
0.65 |
| chr17_30335047_30335869 | 0.18 |
LRRC37B |
leucine rich repeat containing 37B |
451 |
0.82 |
| chr19_39368685_39368907 | 0.18 |
RINL |
Ras and Rab interactor-like |
98 |
0.92 |
| chr16_89626287_89626438 | 0.18 |
RPL13 |
ribosomal protein L13 |
703 |
0.46 |
| chr20_56193926_56194226 | 0.18 |
ZBP1 |
Z-DNA binding protein 1 |
1374 |
0.49 |
| chr10_29970067_29970318 | 0.18 |
ENSG00000222092 |
. |
30652 |
0.18 |
| chr20_49547938_49548164 | 0.18 |
ADNP |
activity-dependent neuroprotector homeobox |
93 |
0.87 |
| chr7_5436621_5436873 | 0.18 |
TNRC18 |
trinucleotide repeat containing 18 |
9087 |
0.16 |
| chr20_1666477_1666686 | 0.18 |
ENSG00000242348 |
. |
19575 |
0.16 |
| chr19_3179098_3179452 | 0.18 |
S1PR4 |
sphingosine-1-phosphate receptor 4 |
539 |
0.66 |
| chr8_145806326_145806590 | 0.18 |
CTD-2517M22.9 |
|
2966 |
0.14 |
| chr12_58176565_58177298 | 0.18 |
TSFM |
Ts translation elongation factor, mitochondrial |
330 |
0.69 |
| chr3_47205224_47205443 | 0.18 |
SETD2 |
SET domain containing 2 |
124 |
0.89 |
| chr12_120427127_120427447 | 0.17 |
CCDC64 |
coiled-coil domain containing 64 |
386 |
0.88 |
| chr4_57843034_57843451 | 0.17 |
POLR2B |
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
646 |
0.6 |
| chr6_24934972_24935145 | 0.17 |
FAM65B |
family with sequence similarity 65, member B |
1130 |
0.57 |
| chr9_37592643_37592824 | 0.17 |
TOMM5 |
translocase of outer mitochondrial membrane 5 homolog (yeast) |
94 |
0.96 |
| chr1_61063097_61063248 | 0.17 |
NFIA |
nuclear factor I/A |
267759 |
0.02 |
| chr12_113495521_113495672 | 0.17 |
DTX1 |
deltex homolog 1 (Drosophila) |
101 |
0.97 |
| chr11_93276609_93276770 | 0.17 |
SMCO4 |
single-pass membrane protein with coiled-coil domains 4 |
15 |
0.98 |
| chr13_19183550_19183860 | 0.17 |
ENSG00000223024 |
. |
259726 |
0.02 |
| chr6_13615615_13616144 | 0.17 |
NOL7 |
nucleolar protein 7, 27kDa |
100 |
0.86 |
| chr11_18127258_18127617 | 0.17 |
SAAL1 |
serum amyloid A-like 1 |
129 |
0.95 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.1 | 0.4 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.2 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.1 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.4 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.1 | 0.2 | GO:0043373 | T-helper cell lineage commitment(GO:0002295) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.1 | 0.2 | GO:0044268 | multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.2 | GO:0019348 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.1 | 0.2 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.1 | 0.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.2 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.2 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.3 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.5 | GO:0045061 | thymic T cell selection(GO:0045061) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.7 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.3 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0032353 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.3 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.1 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0070230 | positive regulation of lymphocyte apoptotic process(GO:0070230) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.0 | GO:0072501 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.3 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.2 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.1 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.3 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.2 | GO:0051000 | positive regulation of nitric-oxide synthase activity(GO:0051000) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.1 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.0 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.1 | GO:0009209 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.2 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0021561 | facial nerve development(GO:0021561) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0045655 | regulation of monocyte differentiation(GO:0045655) |
| 0.0 | 0.1 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.2 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.1 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002823) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.4 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0031060 | regulation of histone methylation(GO:0031060) negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.0 | GO:0046132 | pyrimidine ribonucleoside biosynthetic process(GO:0046132) |
| 0.0 | 0.0 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.0 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 0.0 | GO:0032069 | regulation of nuclease activity(GO:0032069) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.0 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0034311 | diol metabolic process(GO:0034311) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.0 | GO:0000470 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.3 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.1 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.0 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0015851 | nucleobase transport(GO:0015851) |
| 0.0 | 0.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.2 | GO:0061641 | DNA replication-independent nucleosome assembly(GO:0006336) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.4 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.0 | 0.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.1 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.0 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.0 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.2 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.3 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.0 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.6 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0043230 | extracellular organelle(GO:0043230) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.0 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.3 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.3 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.0 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.6 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.7 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.0 | REACTOME PI3K CASCADE | Genes involved in PI3K Cascade |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.0 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 0.0 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.1 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.9 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.3 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |